Ontology Annotator
Contents
Description
The Ontology Annotator (OA) is a curation tool developed by WormBase for a variety of curation purposes including the curation of phenotypes, attaching GO terms to genes, genetic interactions, transgenes, free-text descriptions of genes, and several other data-types. The OA uses CGI, javascript and a postgreSQL database and is web-based, eliminating issues that may arise due to operating system differences and the need for an user to install other dependent software. The OA in many ways is similar to [Phenote] (phenote.org). The OA mainly consists of an Editor, where new data is entered or where pre-existing data can be queried and edited, a Data-Table for data review and a Term-Information panel where information about a term like IDs, synonyms etc., is displayed. The OA includes features like term autocomplete from pre-loaded ontologies, a fast AJAX loading of terms, ability to save/query to/from a postgreSQL database, duplicating data, editing several lines of data at once, and filtering the data. The display to some extent can be custom-organized as columns in the Data-Table can be sorted by dragging and the width of each data-column adjusted. For complex data-types with several fields, the OA allows a tabbed organization.
List of WormBase OA configurations:
- Antibody
- Concise description
- Construct
- Disease
- Disease Term
- Expression pattern
- Gene class
- Gene ontology
- Gene regulation
- Genotype
- Interaction
- Molecule
- Movie
- Phenotype
- Picture
- Process Term
- RNAi
- Topic
- Sequence Feature
- Transgene
The OA uses :
- Perl CGI.
- Yahoo!'s YUI library and a local javascript file.
- PostgreSQL database backend (could probably be modified to other SQL databases).
- Apache webserver.
- Documentation for main CGI, javascript, and modules:OA docs
The above description can also be found here: Web-page for OA
Documentation for existing OAs
OA forms, tables, scripts, etc.
Datatype OAs:
- Antibody
- Concise description
- Construct
- Disease
- Disease Term
- Expression pattern
- Gene class
- Gene ontology
- Gene regulation
- Genotype
- Interaction
- Molecule
- Movie
- Phenotype
- Picture
- Process Term
- RNAi
- Topic
- Sequence Feature
- Transgene
Wish List
- Include dependencies wherever possible. For example, if making an IMP annotation for a given gene, have a gene-specific drop down menu of alleles or RNAi experiments for the WITH column. Or, if making an IGI annotation for the paper, have a drop down list of all genes mentioned in the paepr. Similarly, when entering a GO term, have the ontology (P, F, or C) get entered automatically. (From Curation Interface Meeting)
- Term information window - information should reflect where cursor is placed in the editor window, e.g. Paper should reflect paper info
Adding multiple new rows simultaneously
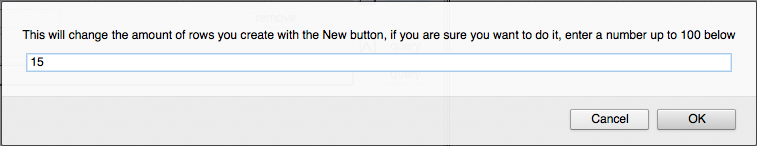
A feature of the OA has been added to allow the addition of many new rows at once. On the left hand side of the "New" button for generating new rows a number is now displayed that indicates the number of new rows that will be created when the "New" button is clicked:
By clicking on the number, curators may specify how many rows they would like to add at once:
Once the number of rows is entered, that number will be displayed to the left of the "New" button:
The curator may then click "New" to add the indicated number of new rows (and hence PGIDs) for that OA.
Batch upload to OA from tab-delimited file
[New as of November 2013] A script has been written that will allow curators to upload data in bulk to the OA through the submission of a properly formatted tab-delimited (TSV) file. The script is located on Mangolassi/Tazendra at:
/home/postgres/public_html/cgi-bin/oa/scripts/populate_oa_tab_file/populate_oa_tab_file.pl
Code Documentation for Batch Upload Script
use strict; use Jex; use LWP::Simple; use URI::Escape; my $domain = 'caltech.edu'; my $path = '~postgres/cgi-bin/oa/ontology_annotator.cgi';
Domain is caltech.edu
The code is accessing the OA to enter data, as opposed to directly entering the data into Postgres tables. This is so that the code can take advantage of the OA-way of entering new rows and handling the different data types.
my $action = '';
my $curator = '';
my $datatype = '';
my $subdomain = '';
my $baseUrl = '';
my %allowedSubdomains;
my @allowedSubdomains = qw( mangolassi tazendra );
foreach (@allowedSubdomains) { $allowedSubdomains{$_}++; }
my $subDomains = join" | ", @allowedSubdomains;
The code above specifically allows for the subdomains of 'mangolassi' or 'tazendra' for entering data. This is implemented at the command line when running the script.
my %allowedDatatypes;
my @allowedDatatypes = qw( app int pro prt rna trp );
foreach (@allowedDatatypes) { $allowedDatatypes{$_}++; }
The code above specifies which OA data type three-letter prefixes are allowed (and hence which OA data types are amenable to batch upload):
- app - Phenotype OA
- int - Interaction OA
- pro - Topic OA
- prt - Process Term OA
- rna - RNAi OA
- trp - Transgene OA
- mop - Molecule OA (added 11/2015)
- cns - Construct OA (added 12/2015)
my %allowedCurators;
my @allowedCurators = qw( 1823 2987 712 );
foreach (@allowedCurators) { $allowedCurators{$_}++; }
≈
The code above specifies which curators (listed as WBPerson ID numbers) can enter data into the OA using this batch upload script.
<pre>
my $usage = qq( ./populate_oa_tab_file.pl <server> <wbpersonID> <filename>\n);
my $printUsage = '';
if ($ARGV[0]) {
if ($allowedSubdomains{$ARGV[0]}) {
$subdomain = $ARGV[0];
$baseUrl = 'http://' . $subdomain . '.' . $domain . '/' . $path; }
else { $printUsage .= qq($ARGV[0] not a valid server, need : $subDomains\n); } }
else { $printUsage .= qq(Need a server : $subDomains\n); }
if ($ARGV[1]) {
my ($num) = $ARGV[1] =~ m/(\d+)/;
if ($allowedCurators{$num}) { $curator = "two$num"; }
else { $printUsage .= qq(WBPerson$ARGV[1] not a valid curator, talk to Juancarlos\n); } }
else { $printUsage .= qq(Need a curator WBPersonID\n); }
unless ($ARGV[2]) { $printUsage .= qq(Need a tab delimited file\n); }
if ($printUsage) { print $usage; print $printUsage; die; }
my $hasError = '';
my $infile = $ARGV[2];
my @lines;
open (IN, "<$infile") or die "Cannot open $infile : $!";
while (my $line = <IN>) {
next if ($line =~ m/^#/);
chomp $line;
push @lines, $line;
} # while (my $line = <IN>)
close (IN) or die "Cannot close $infile : $!";
The code above indicates that it should ignore lines in the input file that start with a hash '#' symbol
my $headerLine = shift @lines;
my (@tables) = split/\t/, $headerLine;
foreach my $table (@tables) {
if ($table =~ m/^([a-z]{3})/) {
if ($allowedDatatypes{$1}) {
if ($datatype) { if ($datatype ne $1) { $hasError .= qq(different datatypes $datatype $1\n); } }
else { $datatype = $1; } }
else { $hasError .= qq($1 not an allowed Datatype\n); }
} # if ($table =~ m/^([a-z]{3})/)
my $url = 'http://' . $subdomain . '.caltech.edu/~postgres/cgi-bin/referenceform.cgi?pgcommand=SELECT+*+FROM+' . $table . '&perpage=1&action=Pg+!';
my $page = get $url;
my ($thereAre) = $page =~ m/There are (\d+) results/;
if ($thereAre == 0) { $hasError .= qq($table has no data in postgres, may not be a valid table\n); }
} # foreach my $table (@tables)
if ($hasError) { print $hasError; die; }
The code above reads in the Postgres table names from the header row of the input file. It reads the first three letters (OA data type three-letter code) from each table name to check if it is a valid OA data type for this script (see above). This code also checks to make sure that each column header corresponds to the same OA data type; if not, the script will produce an error message and stop running. For each table, the code is querying Postgres through the 'referenceform.cgi' to check that there is data in the table, to implicitly check whether the table exists (Note: if a new table is generated without any data, this script will consider the table invalid until data is entered some other way).
my @pgids;
my $newRowAmount = scalar(@lines);
my $url = $baseUrl . '?action=newRow&newRowAmount=' . $newRowAmount . '&datatype=' . $datatype . '&curator_two=' . $curator;
# print "URL $url\n";
my $pageNewLine = get $url;
if ($pageNewLine =~ m/OK\t DIVIDER \t([\d,]+)/) { print "Created pgids $1\n"; (@pgids) = split/,/, $1; }
else { print "Did not get pgid(s) from $url\n"; die; }
The code above adds a new PGID (OA row) for each line in the file (aside from the header row and rows that are commented out) and performs this through the 'newRow' action of the OA for that data type and curator (as if clicking the "New" button on the OA).
my $nonFatalError = '';
foreach my $j (0 .. $#lines) {
my $pgid = $pgids[$j];
unless ($pgid) { $nonFatalError .= qq(No pgid for inputline from array, skipping $lines[$j]\n); next; }
# print "LINE $line\n";
my (@data) = split/\t/, $lines[$j];
for my $i (0 .. $#data) {
my $table = $tables[$i];
unless ($table) { $nonFatalError .= qq(No postgres table for field from array, skipping $tables[$i]\n); next; }
my ($field) = $table =~ m/^${datatype}_(\w+)/;
unless ($field) { $nonFatalError .= qq(No field for postgres table with $datatype, skipping $table\n); next; }
my $data = &convertDisplayToUrlFormat($data[$i]);
my $url = $baseUrl . '?action=updatePostgresTableField&pgid=' . $pgid . '&field=' . $field . '&newValue=' . $data . '&datatype=' . $datatype . '&curator_two=' . $curator;
# print "URL $url\n";
my $pageUpdate = get $url;
unless ($pageUpdate eq 'OK') { $nonFatalError .= qq(Update failed for $pgid changing $table to $data[$i]\n); }
} # for my $i (0 .. $#data)
} # foreach my $line (@lines)
if ($nonFatalError) { print $nonFatalError; }
sub convertDisplayToUrlFormat {
my ($value) = @_;
if ($value) { # if there is a display value replace stuff
($value) = uri_escape($value); # escape all with URI::Escape
# if ($value =~ m/\+/) { $value =~ s/\+/%2B/g; } # replace + with escaped +
# if ($value =~ m/\#/) { $value =~ s/\#/%23/g; } # replace # with escaped #
}
return $value; # return value in format for URL
} # sub convertDisplayToUrlFormat
The code above will check to make sure that each line of the input file was associated with a given PGID number; if there is no PGID for a line (which shouldn't really ever happen) the line will be skipped and a warning message will be written to the screen.
The code checks for a Postgres table name in a given column header and skips data from columns that do not have a Postgres table name in the header row. So, data may be entered without a column header and it will just be skipped by the script.
The code also checks to make sure that there is a valid table name after the three-letter data type code. Valid table names consist of letters, digits, and underscores (no special characters or punctuation). If the table name is not valid, the script will throw an error.
Because the data is being entered through a URL, the text of the data needs to be 'escaped' so as not to interfere with URL syntax.
The code above enters data for each cell/field in the file and performs this through the 'updatePostgresTableField' action of the OA for that data type, field, curator, and PGID.
Usage
Make a symlink of the script in your directory on mangolassi and on tazendra (for example):
$ln -s /home/postgres/public_html/cgi-bin/oa/scripts/populate_oa_tab_file/populate_oa_tab_file.pl* populate_oa_tab_file.pl
scp a testfile.tsv to the same directory on mangolassi and the 'realfile.tsv' to the same directly on tazendra.
Enter (cd into) the directory with the script and run by entering:
./populate_oa_tab_file.pl mangolassi #### testfile.tsv
to enter into the Sandbox OA (on Mangolassi) where '####' is the curator's WBPerson ID (number only), and 'testfile.tsv' is a test upload file to make sure everything is formatted properly
./populate_oa_tab_file.pl tazendra #### realfile.tsv
to enter into the Live OA (on tazendra) where '####' is the curator's WBPerson ID, and 'realfile.tsv' is the real upload file that has been successfully tested on the sandbox (mangolassi)
Note: It is very important to test your batch upload file on the sandbox first, as there are many possible errors in formatting (mis-spelled names, IDs, etc.). Once uploaded to the OA on Tazendra, each entry will need to be edited manually, one-by-one if there are any mistakes.
The script can be run from either mangolassi or tazendra; the important thing is that the destination of the batch upload is specified in the command line when running the script.
Once the script has been run without any errors, the command line should return the list of Postgres IDs (PGIDs) for the new objects created in the relevant OA <note this output might not occur>. For example:
Created pgids 40140,40141,40142
The curator can then paste these IDs, verbatim (comma separated, no spaces), into the PGID field of the associated OA to query for each of the new rows and check the data.
OA's capable of accepting bulk upload
As of November 2013, the list of OA's that can accept bulk uploads via this method are as follows:
- Interaction
- Phenotype
- Process Term
- RNAi
- Topic
- Transgene
- Molecule (added 11/2015)
- Construct (added 12/2015)
Tab-delimited (TSV) file format
It is important that the TSV file be formatted properly. Each column header must be a Postgres table name into which data will be uploaded and match the corresponding OA table entry point such that each row in the spreadsheet/TSV file will be a single entry (with a unique PGID) in the OA/Postgres.
For example: to enter a batch of variation phenotypes, the column headers must point to the phenotype OA app_ tables, the variation and phenotype must match the ID format for the object, not the public names.
app_variation app_paper app_term WBVar00146429 WBPaper00040589 WBPhenotype:0000062 WBVar00146431 WBPaper00040589 WBPhenotype:0000062 WBVar00146432 WBPaper00040589 WBPhenotype:0000062 WBVar00146433 WBPaper00040589 WBPhenotype:0000062 WBVar00146435 WBPaper00040589 WBPhenotype:0000062 WBVar00146437 WBPaper00040589 WBPhenotype:0000062 WBVar00146438 WBPaper00040589 WBPhenotype:0000062 WBVar00146441 WBPaper00040589 WBPhenotype:0000062 WBVar00146444 WBPaper00040589 WBPhenotype:0000062
Note: Every entry in a cell directly below a column header/Postgres table name MUST be entered EXACTLY as can be received by that table/field, i.e. if an 'ontology' or 'multiontology' field, a drop-down menu, or any other controlled vocabulary field, the entries must be formatted appropriately. If these data are not entered in the proper format, the data will enter the Postgres tables (incorrectly) but will not necessarily show up in the OA, making it difficult to track down erroneous data entries.
Any misspelled multi-ontology entries will be ignored by the OA, although the data WILL get written to Postgres; only by querying out the entire object in the OA and making an actual change to the field (by adding or deleting something) will the postgres table be overwritten by what is seen in the OA
Misspelled single ontology fields will show up in the OA (and write to Postgres) AS IS and will need to be fixed manually; note, the OA might still try to recognize the entity, for example if the bogus paper ID "WBPaper00012X45" is entered it will pull up term info for "WBPaper00000012" as a best guess
In order to see how data are typically formatted for a given Postgres table, use the following Postgres query to see sample data entry for a given Postgres table, in this case for the 'rna_phenotype' table:
SELECT * FROM rna_phenotype;
or if you want specifically entries with multiple values:
SELECT * FROM rna_phenotype WHERE rna_phenotype ~ ',';
Google spreadsheet for generating TSV file
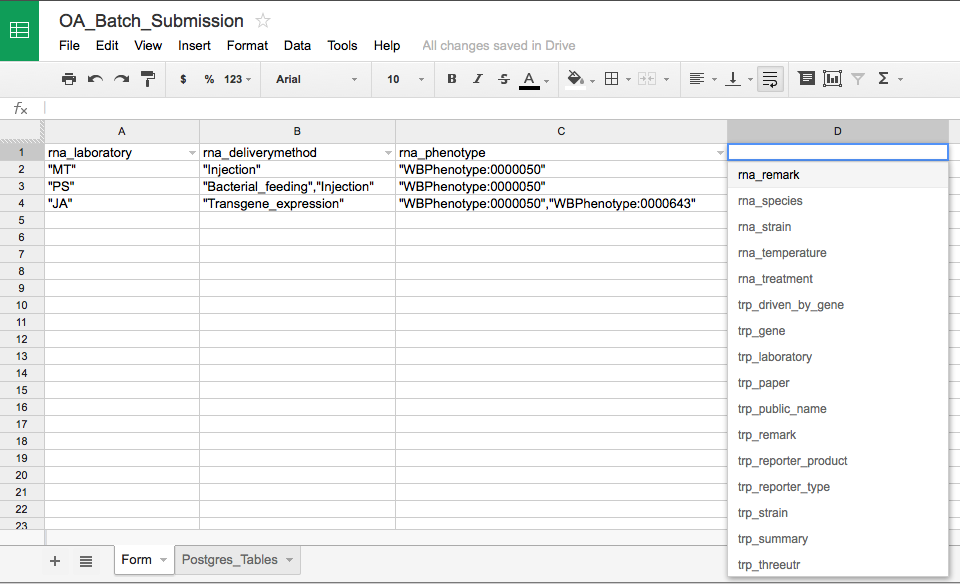
Any tab-delimited (TSV) file should work, but in order to standardize the submission process, a Google spreadsheet has been generated with drop down fields in each header row for selecting the correct Postgres table name:
https://docs.google.com/spreadsheet/ccc?key=0AgaLIpaBTJmSdEVoTXFLdHROOXQ1QzlkZ1VhYVpGMFE#gid=0
Above is a screenshot of the Google spreadsheet form. Note that the Postgres table names that are made available from the drop down menu are stored on the second worksheet of the Google spreadsheet and will need to updated in the event of changes to table names or additions/removals.
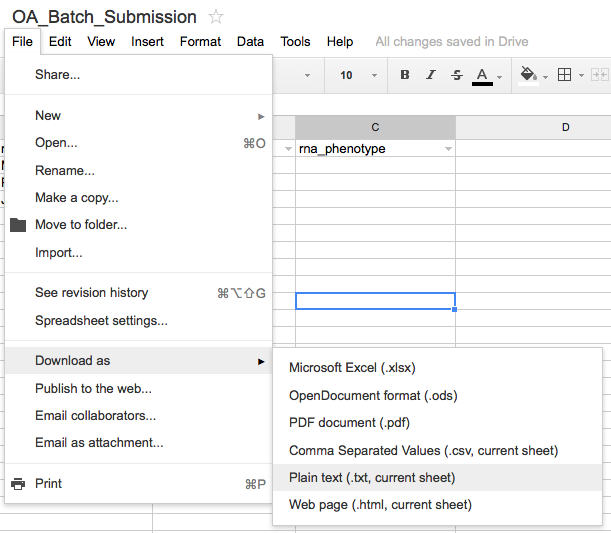
To download the form as a TSV file, select "Download As" from the "File" menu and select "Plain Text".
Even though the file menu suggests that the file will download as a "*.txt" file, it should download the spreadsheet in tab-delimited (TSV) file format which can be directly uploaded to the OA through the batch OA submission script.
Commenting out lines
Any lines in the uploaded TSV file that begin with a hash symbol '#' will be 'commented out' and thus ignored by the script. This may be useful for testing or other purposes. To comment out lines from the Google spreadsheet, just enter a '#' as the first character in the first column on a given row.
Note: Any blank lines submitted in the TSV file will generate empty new rows in the relevant OA
Multiple entries per field/cell
Any fields that can accept multiple entries (e.g. multi-ontology fields or multi-drop-down fields) should have entries entered with double quotes, like
"<entry>"
If multiple entries are being made at once in a single cell/field, enter all entries with double quotes and separated by COMMAS ONLY, NO SPACES, like
"<entry1>","<entry2>","<entry3>"
Large text fields
There are a couple of points to make about large text fields (e.g. Remark):
- Try to avoid entering literal line breaks ('\n') in large text field submissions. The presence of such line breaks may break the code and cause it to crash when running.
- There is no need to enter quotes (single or double) around a large text field entry
- For "DNA Text" fields that accept 'ATGC...' etc., multiple entries ought to be entered with no quotes and separated by a <space><pipe><space> so that they are recognized as distinct entries. So, for example, two DNA text entries should be entered like this:
CAGTATATCGGAGCTGAGTAGCTGATCACAGGACTGTAGCGTCAGT | CTAGTGATGCTTGATCGTATAGTCCGTAC
and NOT like this:
CAGTATATCGGAGCTGAGTAGCTGATCACAGGACTGTAGCGTCAGT|CTAGTGATGCTTGATCGTATAGTCCGTAC
or this:
"CAGTATATCGGAGCTGAGTAGCTGATCACAGGACTGTAGCGTCAGT" | "CTAGTGATGCTTGATCGTATAGTCCGTAC"
Problems
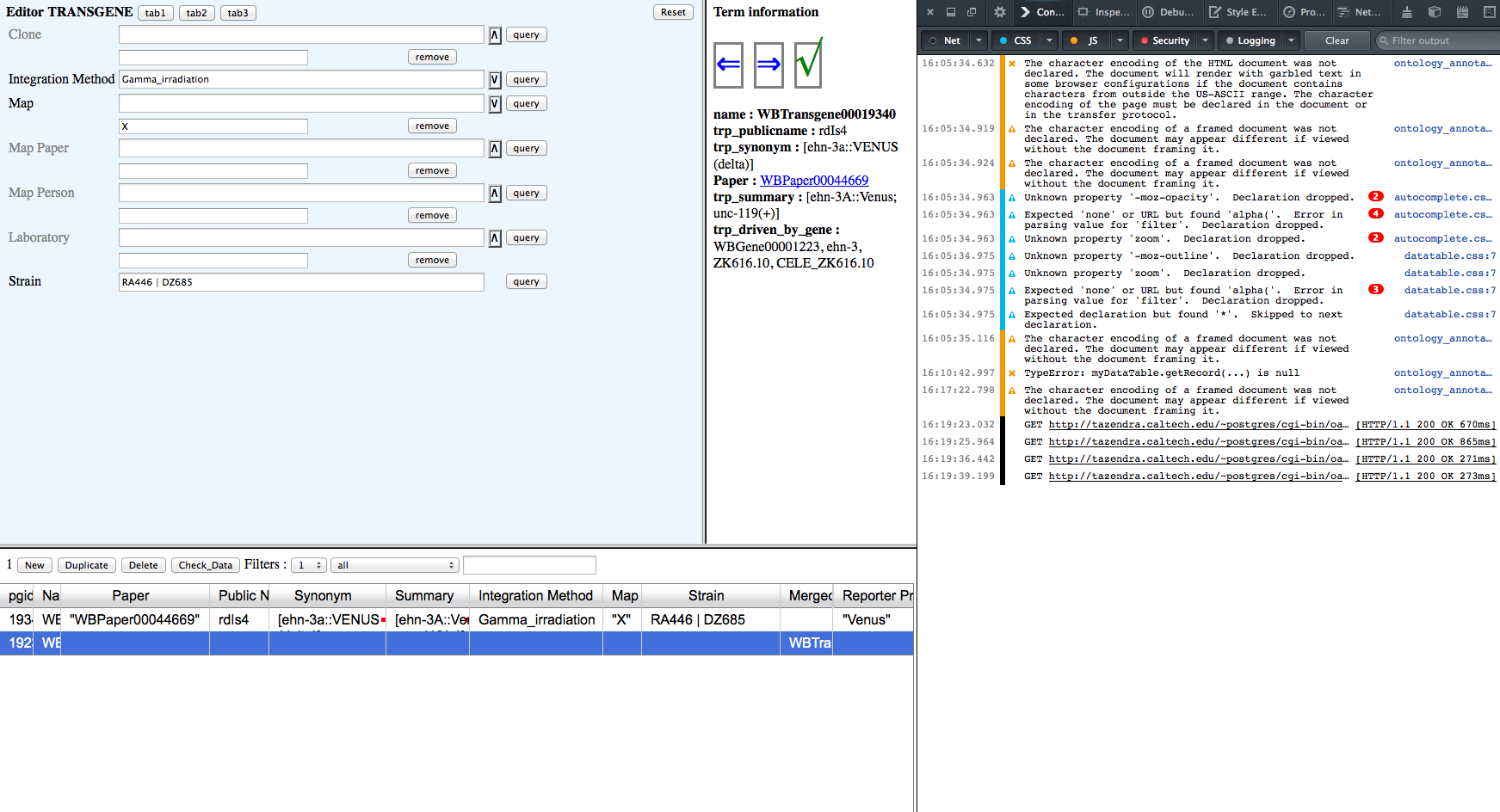
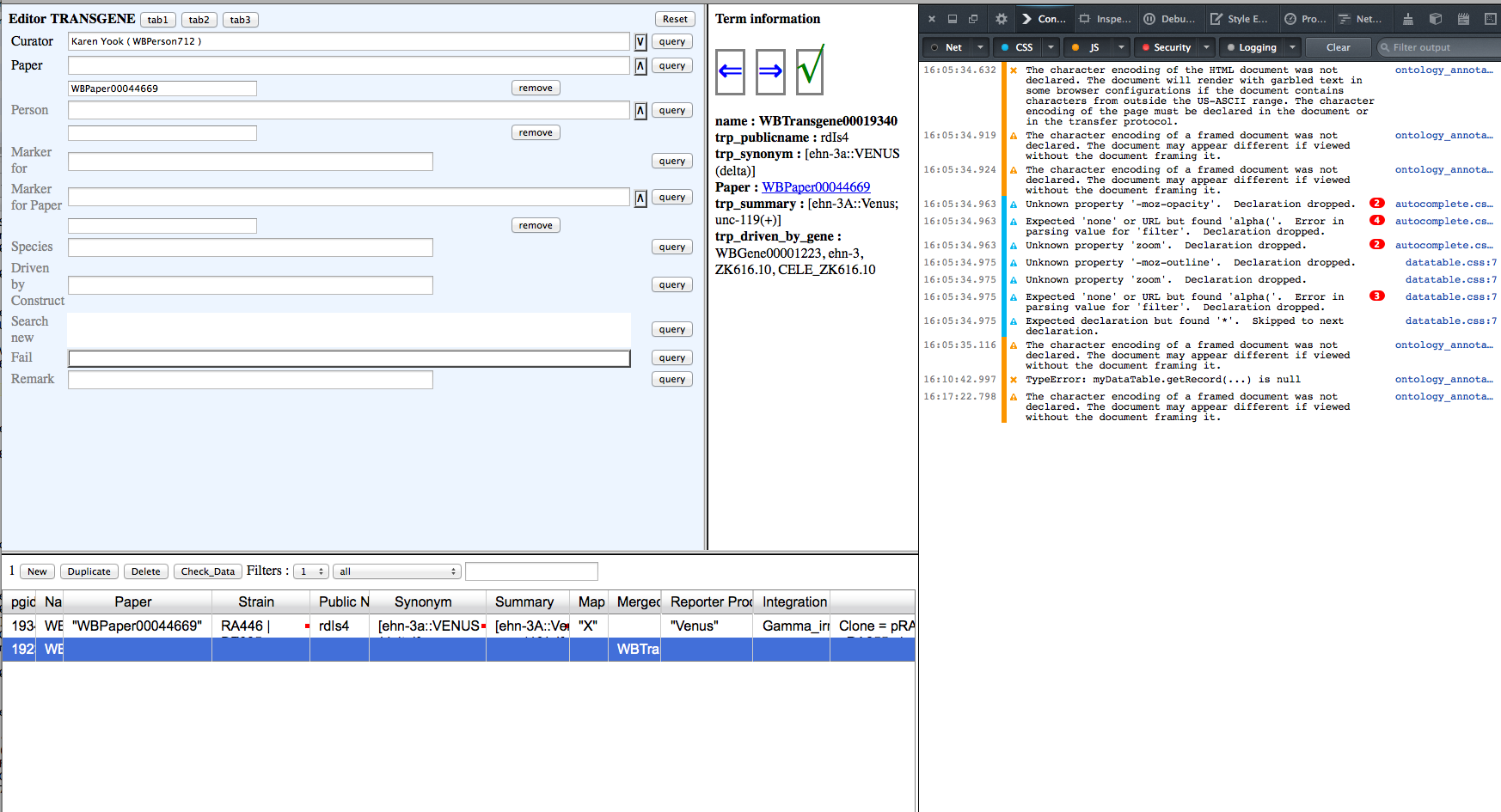
Values in editor window fields persist when they should be blank. The values are correct in the table view, but will be overwritten if an editor field containing information is clicked. see images:
Integration, Map and Strain values are showing in editor window but are blank in postgres. These values should not be in the editor field view for this postgres line, they correctly belong to the top line in the table. I've included the Firefox console in case that can illuminate the bad behavior.
 Back to Caltech documentation
Back to Caltech documentation