RNAi
Archived RNAi documentation may be found here
Contents
RNAi Curation Mission Summary
The term "RNAi" stands for "RNA-interference" and refers to the targeted silencing of gene expression of a "target gene" via introduction of double stranded RNA (dsRNA) containing high degrees of sequence identity to the "target gene". In C. elegans, molecules of dsRNA may be introduced into the worm by a variety of different methods including direct micro-injection, soaking of worms in solution containing dsRNA, feeding worms bacteria that express dsRNA from a plasmid, and transgenic expression (within the worm) of dsRNA. For efficient and specific knockdown of gene expression, the dsRNA must have a minimum sequence identity with the target gene sequence. Any phenotype(s) resulting from the RNAi-mediated knockdown of a particular gene is thought to directly reflect the phenotype of a loss-of-function mutation in that gene, hence providing evidence as to the gene's biological function.
The goal of RNAi curation is to associate RNAi-mediated phenotypes with the target genes knocked down in RNAi experiments, as found in the literature pertaining to C. elegans and related species. There are some important things to consider when curating RNAi experiments. Some RNAi experiments are simple to curate as the strain is N2 (wild type genotype), only a single gene is targeted for RNAi, the phenotype is clearly stated, and the source and identity of the dsRNA is clearly stated by the authors. In many cases, however, the curation task is not so simple: there may be complex genotypes with multiple mutations in the strain receiving the dsRNA, multiple RNAi gene targets, complex genetic interactions, and/or missing descriptions from the authors as to controls, dsRNA delivery method, dsRNA identity, and/or phenotype directly resulting from the RNAi in question. Hopefully these issues will all be addressed below.
RNAi Data Model
This is the ?RNAi data model as of WormBase Release WS251:
//////////////////////////////////////////////////////////////////
//
// ?RNAi class
//
//////////////////////////////////////////////////////////////////
?RNAi Evidence #Evidence
History_name UNIQUE ?Text
Homol Homol_homol ?Homol_data XREF RNAi_homol ?Method Float Int UNIQUE Int Int UNIQUE Int #Homol_info
Sequence_info DNA_text Text UNIQUE Text //stores actual probe sequence for automated mapping
// 1st Text is DNA, 2nd is probe name
Sequence ?Sequence XREF RNAi //links to a real Sequence object used in the experiment
// such as yk clone; not UNIQUE anymore
Clone ?Clone XREF Used_in_RNAi // Chris WS244
PCR_product ?PCR_product XREF RNAi // links to a PCR_product object used in
// the experiment; not UNIQUE anymore
Uniquely_mapped //boolean; if present, signifies that ?RNAi object has a unique sequence
// which maps to a single place in the genome
Experiment Laboratory ?Laboratory
Date UNIQUE DateType
Strain UNIQUE ?Strain
Genotype UNIQUE ?Text //used when no Strain object exists
Treatment UNIQUE ?Text
Life_stage UNIQUE ?Life_stage
Temperature UNIQUE Int
Delivered_by UNIQUE Bacterial_feeding //RL [010327]
Injection //RL [010327]
Soaking //RL [010327]
Transgene_expression //RL [010327]
Inhibits Predicted_gene ?CDS XREF RNAi_result #Evidence // "gene" parent (unreliable)
Gene ?Gene XREF RNAi_result #Evidence //RL [010327]
Transcript ?Transcript XREF RNAi_result #Evidence // [021126 krb]
Pseudogene ?Pseudogene XREF RNAi_result #Evidence // [030801 krb]
Supporting_data Movie ?Movie XREF RNAi // Lincoln, krb [010807]
DB_info Database ?Database ?Database_field ?Accession_number
Species UNIQUE ?Species
Interaction ?Interaction
Reference UNIQUE ?Paper XREF RNAi //[070215 ar2] made reference unique so Paper sort of
// equates to a Study class for Will S
Phenotype ?Phenotype XREF RNAi #Phenotype_info
Phenotype_not_observed ?Phenotype XREF Not_in_RNAi #Phenotype_info
Expr_profile ?Expr_profile XREF RNAi_result // connection added during build [030106 krb]
Remark ?Text #Evidence
Method UNIQUE ?Method
Here is the WS251 ?Phenotype_info model:
////////////////////////////////////////////
//
// ?Phenotype_info Class
//
////////////////////////////////////////////
#Phenotype_info Paper_evidence ?Paper
Person_evidence ?Person
Curator_confirmed ?Person
Remark ?Text #Evidence // specific remarks about the phenotype
Quantity_description ?Text #Evidence //Remark to describe what quantity describes, below
Quantity UNIQUE Int UNIQUE Int #Evidence
Not #Evidence //This is being phased out but is needed for the next phase [06/08/10].
Penetrance Incomplete Text #Evidence
Low Text #Evidence
High Text #Evidence
Complete Text #Evidence
Range UNIQUE Int UNIQUE Int #Evidence // Range of penetrance
Recessive #Evidence
Semi_dominant #Evidence
Dominant #Evidence
Haplo_insufficient #Evidence
Caused_by_gene ?Gene #Evidence
Caused_by_other ?Text #Evidence
Rescued_by_transgene ?Transgene
Variation_effect Gain_of_function_undetermined_type #Evidence
Antimorph_gain_of_function #Evidence
Dominant_negative_gain_of_function #Evidence
Hypermorph_gain_of_function #Evidence
Neomorph_gain_of_function #Evidence
Loss_of_function_undetermined_extent #Evidence
Null #Evidence
Predicted_null_via_sequence #Evidence
Probable_null_via_phenotype #Evidence
Hypomorph_reduction_of_function #Evidence
Predicted_hypomorph_via_sequence #Evidence
Probable_hypomorph_via_phenotype #Evidence
Wild_allele #Evidence
Affected_by Molecule ?Molecule #Evidence // ?Molecule model Karen Yook
EQ_annotations Anatomy_term ?Anatomy_term ?PATO_term #Evidence
Life_stage ?Life_stage ?PATO_term #Evidence
GO_term ?GO_term ?PATO_term #Evidence
Molecule_affected ?Molecule ?PATO_term #Evidence
Temperature_sensitive Heat_sensitive Text #Evidence
Cold_sensitive Text #Evidence
Maternal UNIQUE Strictly_maternal #Evidence
With_maternal_effect #Evidence
Paternal #Evidence
Phenotype_assay Strain ?Strain #Evidence
Treatment ?Text #Evidence
Temperature ?Text #Evidence
Genotype ?Text #Evidence
Ease_of_scoring UNIQUE ES0_Impossible_to_score #Evidence
ES1_Very_hard_to_score #Evidence
ES2_Difficult_to_score #Evidence
ES3_Easy_to_score #Evidence
Here is the WS251 #Evidence hash model:
////////////////////////////////////////////////////////////////////////////////
// Evidence hash
////////////////////////////////////////////////////////////////////////////////
#Evidence Paper_evidence ?Paper // Data from a Paper
Published_as ?Text // .. track other names for the same data
Person_evidence ?Person // Data from a Person
Author_evidence ?Author UNIQUE Text // Data from an Author
Accession_evidence ?Database ?Accession_number // Data from a database (NDB/UNIPROT etc)
Protein_id_evidence ?Text // Reference a protein_ID
GO_term_evidence ?GO_term // Reference a GO_term
Expr_pattern_evidence ?Expr_pattern // Reference a Expression pattern
Microarray_results_evidence ?Microarray_results // Reference a Microarray result
RNAi_evidence ?RNAi // Reference a RNAi knockdown
CGC_data_submission // bless the data as comning from CGC
Curator_confirmed ?Person // bless the data manually
Inferred_automatically Text // bless the data via a script
Date_last_updated UNIQUE DateType // Stores last update timestamp
Feature_evidence ?Feature // Reference a Feature - eg for creation of isoform based on TEC-RED SL2
Laboratory_evidence ?Laboratory // Reference a Lab
From_analysis ?Analysis // Reference an analysis
Variation_evidence ?Variation // Explicitly record variation from which IMP manual GO annotations are made
Mass_spec_evidence ?Mass_spec_peptide
Sequence_evidence ?Sequence // for sequence data that hasn't been submitted to a public resource
Remark ?Text
RNAi annotations in CitaceMinus
These papers have >2000 RNAi objects and, therefore, did not get parsed into postgres tables for RNAi (a total of 59,656 RNAi objects stored in CitaceMinus) :
- WBPaper00004402 (2287 RNAi objects)
- WBPaper00004403 (2584 RNAi objects)
- WBPaper00004651 (2479 RNAi objects)
- WBPaper00005654 (14253 RNAi objects)
- WBPaper00006395 (3230 RNAi objects)
- WBPaper00024497 (10951 RNAi objects)
- WBPaper00025054 (20709 RNAi objects)
- WBPaper00029258 (3163 RNAi objects)
RNAi Curation Standard Operating Procedure (SOP)
In order to ensure consistency of RNAi curation across curators and WormBase releases, a set of standard procedures for RNAi curation are outlined below. The descriptions of these procedures include use of the two main methods for generating *.ace files for release submission: (1) the web-based CGI form for one-at-a-time RNAi object generation and (2) the batch form submission method.
Minimum Requirements for an RNAi Object
Regardless of which curation method you (the curator) choose, there are a set of minimum requirements in order to generate a complete RNAi experiment object:
1) A reference (e.g. WBPaperID)
2) A curator name
3) The sequence of the dsRNA used in the experiment to knockdown gene expression
4) A dsRNA delivery method (e.g. Injection) [Note: some authors omit this information. This can be curated arbitrarily and removed from the *.ace file once it has been generated]
5) A phenotype (observed or not_observed)
If any one of these five basic pieces of information are missing, the scripts that generate the *.ace file will fail, returning an error.
RNAi Paper Flagging and Processing of SVM "Low" Results and Tracking
We have been using SVM to flag papers that may contain RNAi data. These papers are given a probability score (High, Medium or Low) based on the SVM training set to reflect the possibility that they contain RNAi data to be curated. The High and Medium scoring papers are automatically added to the list of RNAi papers that are to be curated (on the Paper Editor) and the Lows are stored separately. The Low scoring papers must be manually checked by a curator and if the paper has "curatable" RNAi data, the curator must manually add that paper to the list. The Low scoring paper list is being checked approximately every 3 months (it is added to automatically every Monday at 2am based on SVM results). The following is the SOP for checking these papers and adding them to the list of RNAi papers to curate.
SVM Low's - SOP for checking papers
The file containing the papers with a low priority score (named "low") is stored on tazendra in the directory: /home/postgres/work/pgpopulation/svm/gary_rnai New papers are concatenated to this list. The curator checking the recently added papers must go back in the list to the most recent paper that has a "commented out" mark "//". This mark means that the list has been checked up to this point in the paper list....all papers after this mark need to be manually checked. When the curator is finished, they should "comment out" the last paper on the list so the next curator knows where to begin checking next time.
SVM Low's - SOP for adding papers to curation pipeline
After checking the "Low" papers, the ones that do conatin data must be added to the curation list. To do so go to the curator_first_pass.cgi on the web which is listed under "curation Forms" on the site map for tazendra. Add the appropriate number of the WBPaper id, for example just "00038308" without the WBPaper prefix, and click on the query button. Under "rnai" in the Gene Function section add the following comment in the curator box "SVM -LOW". When you hit "Flag" at the bottom of the page this paper will be added to the RNAi curation list and noted as an SVM with a low probability score.
SVM Low's - SOP for adding papers SVM Tracking
For tracking of SVM results (if False Positive, False Neg etc) a tool was created: http://XXXXXXXXX.caltech.edu/~postgres/cgi-bin/svm_results.cgi This web-based tool will automatically track papers that are positive based on our RNAi curation pipeline/checkout form and the OA; That is if a paper is on the checkout form and is curated, the data will be in the OA and marked as a True Positive. If the paper on the checkout list is a False Positive, the curator must note this on the paper editor (Flag False Positive) and the results will be forwarded to the SVM tracking form. http://tazendra.caltech.edu/~postgres/cgi-bin/paper_editor.cgi The SVM lows that do contain RNAi data are added to the checkout form by a curator (see above) and will be dealt with as any other paper on the list. The SVM lows that actually do not contain RNAi data must be entered manually onto the SVM tracking form as False Positives. To do this go to http://XXXXXXXXX.caltech.edu/~postgres/cgi-bin/svm_results.cgi Under "Enter Curator Results" Select data type/ select curator negative / add the list of negatives to / select comment "SVM positive - Curator negative"
RNAi OA Postgres Tables (rna_*)
CG updated 9-1-2015
| postgres tables | ||
| rna_anatomy | rna_goprocess | rna_person_hst |
| rna_anatomyquality | rna_goprocessquality | rna_phenotype |
| rna_child_of | rna_heatsens | rna_phenotype_hst |
| rna_child_of_hst | rna_heatsens_hst | rna_phenotypenot |
| rna_coldsens | rna_historyname | rna_phenotypenot_hst |
| rna_coldsens_hst | rna_historyname_hst | rna_phenremark |
| rna_curator | rna_laboratory | rna_phenremark_hst |
| rna_curator_hst | rna_laboratory_hst | rna_quantdesc |
| rna_database | rna_lifestage | rna_quantdesc_hst |
| rna_database_hst | rna_lifestage_hst | rna_quantfromto |
| rna_date | rna_lifestagequality | rna_quantfromto_hst |
| rna_date_hst | rna_molaffected | rna_remark |
| rna_deliverymethod | rna_molaffectedquality | rna_remark_hst |
| rna_deliverymethod_hst | rna_molecule | rna_sequence |
| rna_dnatext | rna_molecule_hst | rna_sequence_hst |
| rna_dnatext_hst | rna_movie | rna_species |
| rna_exprprofile | rna_movie_hst | rna_species_hst |
| rna_exprprofile_hst | rna_name | rna_strain |
| rna_flaggenereg | rna_name_hst | rna_strain_hst |
| rna_flaggenereg_hst | rna_nodump | rna_suggested |
| rna_flaggeneticintxn | rna_nodump_hst | rna_suggested_definition |
| rna_flaggeneticintxn_hst | rna_paper | rna_suggested_definition_hst |
| rna_fromgenereg | rna_paper_hst | rna_suggested_hst |
| rna_fromgenereg_hst | rna_pcrproduct | rna_temperature |
| rna_genotype | rna_pcrproduct_hst | rna_temperature_hst |
| rna_genotype_hst | rna_penetrance | rna_treatment |
| rna_gocomponent | rna_penetrance_hst | rna_treatment_hst |
| rna_gocomponentquality | rna_penfromto | |
| rna_gofunction | rna_penfromto_hst | |
| rna_gofunctionquality | rna_person |
RNAi OA
CG updated 9-1-2015
NOTE: Warning: RNAi objects may have multiple lines in the OA due to the nested phenotype data (Penetrance info, etc.)
Dependency Notice: The Community Phenotype Form and the RNAi OA each have separate code to look up the most recent RNAi ID and generate new RNAi IDs based on that. If the code is changed for one, we should also check that the code is changed for the other.
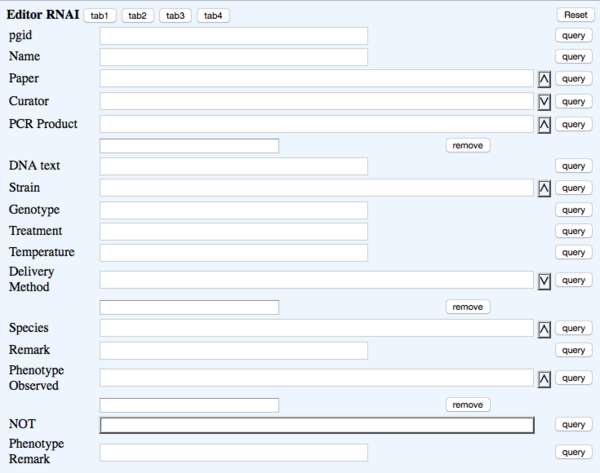
TAB 1
- pgid - the postgres ID - NOT DUMPED
- Name - rna_name - RNAi : - WBRNAiID #Note: This is automatically assigned
- Paper - rna_paper - Reference - ?Paper (Ontology)
- Curator - rna_curator - NOT DUMPED - Curator (Dropdown)
- PCR Product - rna_pcrproduct - PCR_product - ?PCR_product (Multi-ontology) #Note: EBI/Hinxton will map these
- DNA Text - rna_dnatext - DNA_text - Big text #Note: multiple Sequences should be separated by pipe "|" (caution: pressing <ENTER> may cause problems)
- Strain - rna_strain - Strain - ?Strain (Ontology)
- Genotype - rna_genotype - Genotype - Big text
- Treatment - rna_treatment - Treatment - Big text
- Temperature - rna_temperature - Temperature - Text (Integer)
- Delivery Method - rna_deliverymethod - Delivered_by - (Multi-dropdown) four choices: "Bacterial_feeding", "Injection", "Soaking", "Transgene_expression"
- Species - rna_species - Species - ?Species (Dropdown)
- Remark - rna_remark - Remark - Big text
- Phenotype Observed - rna_phenotype - Phenotype - ?Phenotype ID & name (Multi-ontology)
- NOT - rna_phenotypenot - Phenotype_not_observed - NOT Toggle
- Phenotype Remark - rna_phenremark - (in-line with Phenotype) Remark - (Big Text)
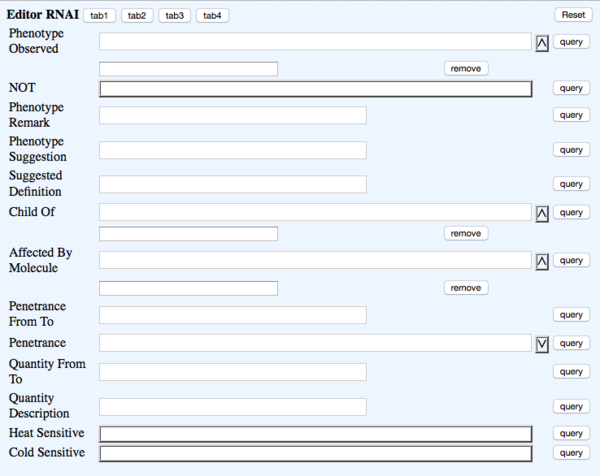
TAB 2
- Phenotype Observed - rna_phenotype - Phenotype - ?Phenotype ID & name (Multi-ontology)
- NOT - rna_phenotypenot - Phenotype_not_observed - NOT Toggle
- Phenotype Remark - rna_phenremark - (in-line with Phenotype) Remark - (Big Text)
- Phenotype Suggestion - rna_suggested - not dumped - field to suggest new phenotype term. Will replace whatever term(s) are currently in the Phenotype field once it is approved
- Suggested Definition - rna_suggested_definition - NOT DUMPED - Big text field with definition of new suggested phenotype term
- Child Of - rna_child_of - NOT DUMPED - list of parent phenotype term(s) for suggested phenotype
- Affected By Molecule - rna_molecule - (in-line with Phenotype) Molecule - ?Molecule (Multi-ontology)
- Penetrance From To - rna_penfromto - (in-line with Phenotype) Range - Text (Integer-space-Integer)
- Penetrance - rna_penetrance - (in-line with Phenotype) Penetrance - dropdown like in phenotype OA
- Quantity From To - rna_quantfromto - (in-line with Phenotype) Quantity - Text (Integer-space-Integer)
- Quantity Description - rna_quantdesc - (in-line with Phenotype) Quantity_description - Big text
- Heat Sensitive - rna_heatsens - (in-line with Phenotype) Temperature_sensitive - Toggle
- Cold Sensitive - rna_coldsens - (in-line with Phenotype) Temperature_sensitive - Toggle
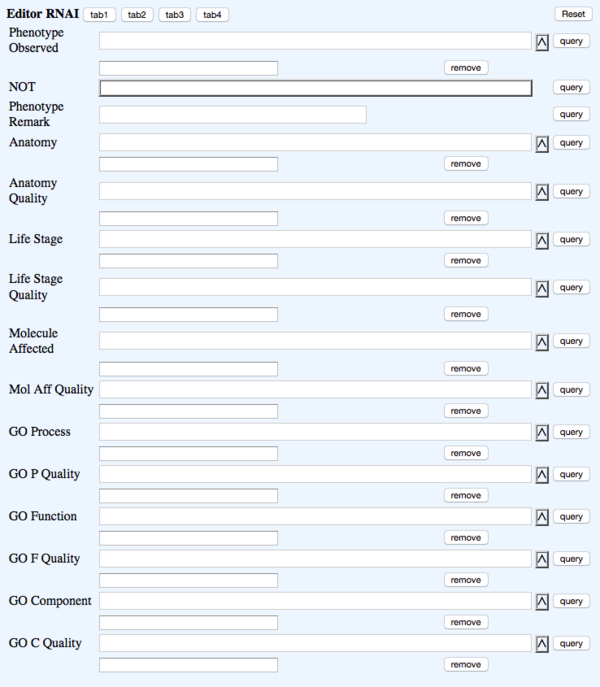
TAB 3
- Phenotype Observed - rna_phenotype - Phenotype - ?Phenotype ID & name (Multi-ontology)
- NOT - rna_phenotypenot - Phenotype_not_observed - NOT Toggle
- Phenotype Remark - rna_phenremark - (in-line with Phenotype) Remark - (Big Text)
- Anatomy - rna_anatomy (Multi-ontology)
- Anatomy Quality - rna_anatomyquality (Multi-ontology)
- Life Stage - rna_lifestage - Life_stage - ?Life_stage (Multi-ontology)
- Life Stage Quality - rna_lifestagequality (Multi-ontology)
- Molecule Affected - rna_molaffected (Multi-ontology)
- Mol Aff Quality - rna_molaffectedquality (Multi-ontology)
- GO Process - rna_goprocess (Multi-ontology)
- GO P Quality - rna_goprocessquality (Multi-ontology)
- GO Function - rna_gofunction (Multi-ontology)
- GO F Quality - rna_gofunctionquality (Multi-ontology)
- GO Component - rna_gocomponent (Multi-ontology)
- GO C Quality - rna_gocomponentquality (Multi-ontology)
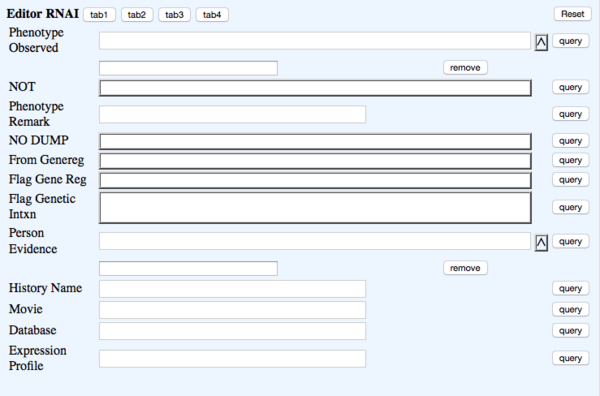
TAB 4
- Phenotype Observed - rna_phenotype - Phenotype - ?Phenotype ID & name (Multi-ontology)
- NOT - rna_phenotypenot - Phenotype_not_observed - NOT Toggle
- Phenotype Remark - rna_phenremark - (in-line with Phenotype) Remark - (Big Text)
- NO DUMP - rna_nodump - NOT DUMPED - toggle - Prevents data from that row from being dumped. This will also indicate to the Curation Status Form (CSF) that the paper has not been curated and will come up as "oa_blank" in the CSF
- From Genereg - rna_fromgenereg - NOT DUMPED - Toggle
- Flag Gene Reg - rna_flaggenereg - NOT DUMPED - Toggle
- Flag Genetic Intxn - rna_flaggeneticintxn - NOT DUMPED - Toggle
- Person Evidence - rna_person - Evidence Person_evidence - multiontology - for ?RNAi object's #Evidence
- History Name - rna_historyname - History_name - Text #Note: this field is to accommodate older RNAi objects
- Movie - rna_movie - Movie - bigtext #Note: this field is to accommodate older RNAi objects; Note: Separate multiple movie entries with bars (|)
- Database - rna_database - Database - Text #Note: this field is to accommodate older RNAi objects; Enter data in this format, split multiple database lines with pipes if there are any :
- Phenobank2 Gene&RNAID GeneID=507328 | Phenobank3 Gene&RNAID GeneID=123456
- Expression Profile - rna_exprprofile - Expr_profile - Text #Note: this field is to accommodate older RNAi objects
NOTES ON SUGGESTING NEW PHENOTYPE TERMS THROUGH THE RNAI OA
- When suggesting a new phenotype term in TAB 2 of the RNAi OA, curators should make sure to create a PGID/Row with a single placeholder phenotype that is intended to be replaced by the new phenotype term once the new term has been approved
- If the data is dumped from the OA for upload before the new term has been approved, the object will dump with the placeholder phenotype term for that upload
- This behavior is similar to that of the Phenotype OA
- The data entered into the three fields "Phenotype Suggestion", "Suggested Definition", and/or "Child Of" will immediately be populated into the corresponding fields in the New Objects CGI
- Cron job:
0 3 * * sun /home/acedb/gary/phn_suggested/phn_suggested_oa.pl
Runs every Sunday at 3 am, checking for entries in rna_suggested (and app_suggested) within the last 7 days and sends Gary S. an e-mail if there are any new entries. If there aren't any, there is no email.
NOT USING
- Evidence - postgres table and field not created, don't know what to parse into this field - Text #Note: this field is to accommodate older RNAi objects So there are only 8 RNAi objects that use the Evidence field, and they do so with 'Person_evidence'. In the OA, I think we should include: Person_evidence, Author_evidence, Curator_confirmed, Laboratory_evidence -- C We're only keeping Person_evidence for the #Evidence hash associated with the whole ?RNAi object -- C+J
- Penetrance Incomplete - rna_penincomplete - Toggle I think we talked about this, but in case we didn't phenotype OA uses a dropdown for the 4 penetrance types instead of separate toggle fields -- J OK, this sounds like a good idea. Let's just make it a dropdown list here as well -- C
- Penetrance Low - rna_penlow - Toggle
- Penetrance High - rna_penhigh - Toggle
- Penetrance Complete - rna_pencomplete - Toggle
- Method - rna_method - Method - Text
Notes
Fields for the tags "Predicted_gene", "Gene", "Transcript", "Pseudogene", "Homol_homol" and "Uniquely_mapped" will be omitted from the OA and populated in ACEDB after probe mapping to the genome at the EBI during the build process.
*.ACE Dumper Documentation
Each RNAi object will need to get dumped with "RNAi" in the "Method" tag
On tazendra, perl module is /home/postgres/work/citace_upload/rnai/get_rnai_ace.pm
Run with script /home/postgres/work/citace_upload/rnai/use_package.pl
Generates rnai.ace.<date> and err.out.<date> (to the day, so it will overwrite previous dumps from the same date) please check dumper and .ace file dumped -- J The dumper seems to be working OK since the dumped *.ACE file you generated is reading in OK, except for the "Evidence" line, as I comment about above in the "Tab3" section. The line currently dumps out as "Person_evidence ..." and it needs to dump out as "Evidence Person_evidence ...", otherwise the file throws errors when reading into ACEDB -- C Fixed, I thought only the innermost tag mattered, but I guess because there's no data in the RNAi part, just the tag, it needs to be there before the #Evidence part -- J I believe so, yes. -- C
Use_package.pl line comments:
'my $outfile = 'rnai.ace.' . $date;' 'my $errfile = 'err.out.' . $date;'
- These lines (above) specify the output; change as necessary.
'my ($all_entry, $long_text, $err_text) = &getRnai('all');'
'# my ($all_entry, $long_text, $err_text) = &getRnai('WBRNAi00008227');'
- The lines above specify what to dump out of the OA. 'all' dumps everything. If you want to dump specific objects, comment out the top line (add a '#') and uncomment the bottom line (remove '#') and specify the object name where 'WBRNAi00008227' is. Because of permissions, the script will need to be copied, modified, and then run from a permissible directory.
Molecule now converts the pgid stored in postgres with the molecule name in mop_name for that pgid -- J
Error Checks During Dump Process
The following is a list of checks that the .ACE dumper script will perform on all RNAi objects being dumped out of the OA to make sure that the data is consistent and doesn't have any nonsensical information. Any errors that are found will be printed to an "err" file. Every line of the file will list an error message of the general format:
PGID <TAB> Dump_status <TAB> Curator ID <TAB> Explanation
where "Dump_status" could be "nodump" (for objects that have a 'fatal' error and will not be dumped) or "flagonly" (for objects that have a 'non-fatal' error and will be dumped).
Fatal Errors (RNAi objects will not get dumped)
1) If there is no identifying RNAi probe information (i.e. the sequence or clone used to knockdown the target gene) in an RNAi object, the dumper script will generate an error message that is printed to the ERROR output file and the object will not get dumped. This is determined by checking that:
a) There is at least one "PCR Product" entry OR
b) There is at least one "DNA Text" entry OR
c) There is at least one "Sequence" entry
If none of these conditions hold true, then an error message will be printed in tab-delimited format like this:
12345 nodump WBPerson1234 There is no sequence, neither pcrproduct nor dnatext nor sequence
2) If there is no reference (Paper or Person) then the object will not get dumped and an error message is printed:
12345 nodump WBPerson1234 There is no reference, neither paper nor person
3) If there is no "Phenotype" specified by the curator, the object will not get dumped and an error message will print to the ERROR output file like this:
12345 nodump WBPerson1234 There is no phenotype
4) If there is no RNAi ID, the object will not get dumped and an error message will print to the ERROR output file like this:
12345 nodump WBPerson1234 There is no RNAi ID
Non-Fatal Errors (RNAi objects will get dumped, but error message will get printed)
1) If there is no dsRNA "Delivery Method" that has been specified by the curator, the RNAi object will get dumped to the .ACE file, but an error message will print to the ERROR output file like this:
12345 flagonly WBPerson1234 There is no deliverymethod
2) If there is no "Species" specified by the curator, the RNAi object will get dumped to the .ACE file, but an error message will print to the ERROR output file like this:
12345 flagonly WBPerson1234 There is no species
3) If no curator is listed for the RNAi object, the RNAi object will get dumped, but an error message will print to the ERROR output file like this:
12345 flagonly no curator has no curator