WormBase Genomes
Contents
WormBase Genomes
This is a record of the current and proposed set of genomes in WormBase.
I think that this page is a correct statement of our intentions.
We may, of course, alter our plans for which species to include as circumstances dictate and so the list of organisms which should be included should be treated as somewhat tentative.
Tiers
The different genomes in WormBase are classified in various tiers which depend on the amount of curation effort we are able to put into maintaining them.
Tier I - All efforts are made to curate the gene structures and any other genetic or metabolic information. Only C. elegans is in this group.
Tier II - Efforts are made, where practical, to manually curate the gene structure and possibly some other genomic information. WormBase 'owns' the assembly in the ENA and GenBank so that new gene annotations can be submitted to the ENA/GenBank by WormBase.
Tier III - We will set up the genome on WormBase with any gene structures that the authors of this genome have predicted. No further curation efforts are made by the WormBase consortium.
Tier IV - Proposed tier for organisms with only transcriptome information and no coherent genome. There are no examples of this in WormBase at present.
Tier V - Proposed tier for organisms with only genome information and no coherent transcriptome. There are no examples of this in WormBase at present.
Genome
The Genome column in the table indicates whether the genome has been added to WormBase.
Gene
The Genes column in the table indicates whether gene structures have been added to WormBase.
The genome status
| Species | NCBI TaxonID | Tier | Genome | Genes | Origin | Comments |
|---|---|---|---|---|---|---|
| Caenorhabditis briggsae | 6238 | II | Yes | Yes | WashU | [Sept 2010] New assembly from Erich Haag being worked on. [Feb 2011] updated in WS224 |
| Caenorhabditis remanei | 31234 | II | Yes | Yes | WashU | |
| Caenorhabditis brenneri (Species 4) |
135651 | II | Yes | Yes | WashU | [Jan 2011] The current assembly contains quite a bit of heterozygosity. Warning WS223-WS226 genome assembly is not in sync with the annotation files or INSDC. Please use WS227+ |
| Caenorhabditis elegans strain N2 | 6239 | I | Yes | Yes | WashU/Sanger | |
| Caenorhabditis elegans strain DR1035 | 6239 | III | Mark Blaxter | status unclear | ||
| Caenorhabditis japonica | 281687 | II | Yes | Yes | WashU | [Jan 2011] New/improved assembly is being worked on at WashU [Oct 2011] new assembly in WS227 |
| Caenorhabditis drosophilae | 96641 | III | WashU | [Sept 2010] being assembled | ||
| Caenorhabditis angaria (species 3 strain PS1010) | 96668 | III | Yes | Yes | CalTech | Added to WormBase in release WS218. [Jan 2011] This species now has an official name of C. angaria |
| Caenorhabditis species 7 strain JU1286 | 870436 | V | WashU | released in WS226. WARNING the genome sequence contains contaminations | ||
| Caenorhabditis species 9 strain JU1422 | 870437 | V | WashU | released in WS226. WARNING the genome sequence contains contaminations | ||
| Caenorhabditis species 11 strain JU1373 | 886184 | III | Yes | Yes | WashU | genome and deNovo gene set released in WS226. Replaced by RNAseq based gene set in WS227. |
| Heterorhabditis bacteriophora | 37862 | V | Yes | WashU | [Jan 2011] Submitted to GenBank - Accession: EF043402 [Sep 2011] Gene set and Annotations are being worked on. [Nov 2011] on WormBase as of WS229 | |
| Pristionchus pacificus | 54126 | II | Yes | Yes | WashU/MPI | [16 December 2010] Updated to the newest assembly and geneset in WS221 |
| Haemonchus contortus | 6289 | III | Yes | Yes | Sanger | Added to WormBase in release WS209. |
| Strongyloides ratti | 34506 | III | Yes | Sanger | [Jan 2011] draft assembly in GenBank, released in WS226 | |
| Meloidogyne hapla | 6305 | III | Yes | Yes | NCSU hapla.org | Added to WormBase in release WS204. |
| Meloidogyne incognita | 6306 | III | Yes | INRA | Added to WormBase in release WS205. Genes are not yet available. | |
| Brugia malayi | 6279 | III | Yes | Yes | TIGR -> WashU/Sanger | [Sept 2010] Currently using the old TIGR assembly. Waiting for WashU (did assembly) and Sanger (did gene models) to publish, then we will use the new assembly. [Dec 2010] merged Augustus gene predictions from Erich Schwarz into WS216 |
| Onchocerca volvulus | 6282 | III | Sanger | |||
| Ascaris suum | 6253 | III | Yes | Yes | Davis | [Oct 2011] integrated the Davis genome without a reference gene set. [Nov 2011] added a reference gene set |
| Globodera pallida | 36090 | III | Sanger | |||
| Nippostrongylus brasiliensis | 27835 | III | Sanger | |||
| Strongyloides ransomi | 553534 | III | Sanger | |||
| Teladorsagia circumcincta | 45464 | III | Sanger | |||
| Trichuris muris | 70415 | III | Sanger | |||
| Trichinella spiralis | 6334 | III | Yes | Yes | WashU | [Sept 2010] Being assembled. [Feb 2011] published in Nature. [Mar 2011] added to WormBase in WS225 |
| Bursaphelenchus xylophilus | 6326 | III | Yes | Yes | Sanger | [Sept 2011] published in PLOS Pathogens [Nov 2011] added to WormBase in WS229 |
| Steinernema carpocapsae | 34508 | Laboratorio Nacional de Genómica para la Biodiversidad / CalTech | [May 2011] Being assembled. |
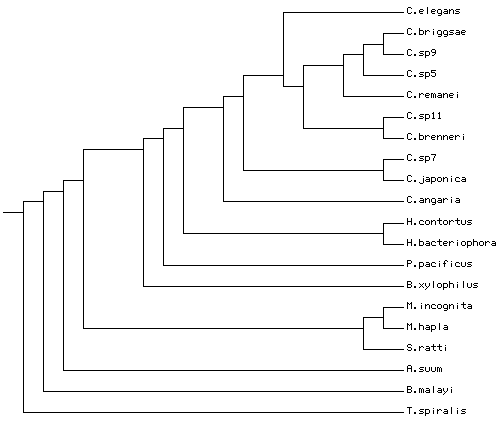
Phylogeny
Given my understanding of the current phylogenetic literature (and based on personal communications with Karin Kiontke), the correct guide tree would be:
((((((((((((C.briggsae,C.sp9),C.sp5),C.remanei),(C.sp11,C.brenneri)),C.elegans),(C.sp7,C.japonica)),C.angaria),(H.contortus,H.bacteriophora)),P.pacificus),((M.incognita,M.hapla), (S.ratti,B.xylophilus))),(A.suum,B.malayi)),T.spiralis);