RNAi
Contents
RNAi Curation Mission Summary
The term "RNAi" stands for "RNA-interference" and refers to the targeted silencing of gene expression of a "target gene" via introduction of double stranded RNA (dsRNA) containing high degrees of sequence identity to the "target gene". In C. elegans, sequences of dsRNA may be introduced into the worm by a variety of different methods including direct micro-injection, soaking of worms in solution containing dsRNA, feeding worms bacteria that express dsRNA from a plasmid, and transgenic expression (within the worm) of dsRNA. For efficient and specific knockdown of gene expression, the dsRNA must have a minimum sequence identity with the target gene sequence. Any phenotype(s) resulting from the RNAi-mediated knockdown of a particular gene is thought to directly reflect the phenotype of a loss-of-function mutation in that gene, hence providing evidence as to the gene's biological function.
The goal of RNAi curation is to associate RNAi-mediated phenotypes with the target genes knocked down in RNAi experiments, as found in the literature pertaining to C. elegans and related species.
RNAi Data Model
This is the RNAi data model as of WormBase Release WS222:
//////////////////////////////////////////////////////////////////
//
// ?RNAi class
//
//////////////////////////////////////////////////////////////////
?RNAi Evidence #Evidence
History_name UNIQUE ?Text
Homol Homol_homol ?Homol_data XREF RNAi_homol ?Method Float Int UNIQUE Int Int UNIQUE Int #Homol_info
Sequence_info DNA_text Text UNIQUE Text //stores actual probe sequence for automated mapping
// 1st Text is DNA, 2nd is probe name
Sequence ?Sequence XREF RNAi //links to a real Sequence object used in the experiment
// such as yk clone; not UNIQUE anymore
PCR_product ?PCR_product XREF RNAi // links to a PCR_product object used in
// the experiment; not UNIQUE anymore
Uniquely_mapped //boolean; if present, signifies that ?RNAi object has a unique sequence
// which maps to a single place in the genome
Experiment Laboratory ?Laboratory
Author ?Author
Date UNIQUE DateType
Strain UNIQUE ?Strain
Genotype UNIQUE ?Text //used when no Strain object exists
Treatment UNIQUE ?Text
Life_stage UNIQUE ?Life_stage
Temperature UNIQUE Int
Delivered_by UNIQUE Bacterial_feeding //RL [010327]
Injection //RL [010327]
Soaking //RL [010327]
Transgene_expression //RL [010327]
Inhibits Predicted_gene ?CDS XREF RNAi_result #Evidence // "gene" parent (unreliable)
Gene ?Gene XREF RNAi_result #Evidence //RL [010327]
Transcript ?Transcript XREF RNAi_result #Evidence // [021126 krb]
Pseudogene ?Pseudogene XREF RNAi_result #Evidence // [030801 krb]
Supporting_data Movie ?Movie XREF RNAi // Lincoln, krb [010807]
Picture ?Picture XREF RNAi
DB_info Database ?Database ?Database_field ?Accession_number //to link out to Phenobank ar2 02-DEC-05 //removed UNIQUE as reqs multiple connections
Species UNIQUE ?Species
Gene_regulation ?Gene_regulation XREF RNAi // this tag is used when an RNAi experiment describes gene regulation ar2 29-MAR-06 for igor
Interaction ?Interaction
Reference UNIQUE ?Paper XREF RNAi //[070215 ar2] made reference unique so Paper sort of equates to a Study class for Will S
Phenotype ?Phenotype XREF RNAi #Phenotype_info
Phenotype_not_observed ?Phenotype XREF Not_in_RNAi #Phenotype_info //added by Wen to separate Not phenotype from real phenotypes
Expr_profile ?Expr_profile XREF RNAi_result // connection added during build [030106 krb]
Remark ?Text #Evidence
Method UNIQUE ?Method
RNAi Curation Standard Operating Procedure (SOP)
In order to ensure consistency of RNAi curation across curators and WormBase releases, a set of standard procedures for RNAi curation are outlined below. The descriptions of these procedures include use of the two main methods for generating *.ace files for release submission: (1) the web-based CGI form for one-at-a-time RNAi object generation and (2) the batch form submission method.
Minimum Requirements for an RNAi Object
Regardless of which curation method you (the curator) choose, there are a set of minimum requirements in order to generate a complete RNAi experiment object:
1) A reference (WBPaperID)
2) A curator name
3) The sequence of the dsRNA used in the experiment to knockdown gene expression
4) A phenotype (observed or not_observed)
Without any one of these four basic pieces of information, the scripts that generate the *.ace file will fail, returning an error.
Two Methods of Curation: Web CGI & Batch Form
Web-based CGI
The web-based RNAi curation form can be found at:
http://elbrus.caltech.edu/cgi-bin/igor/rnaitools/rnai_curation
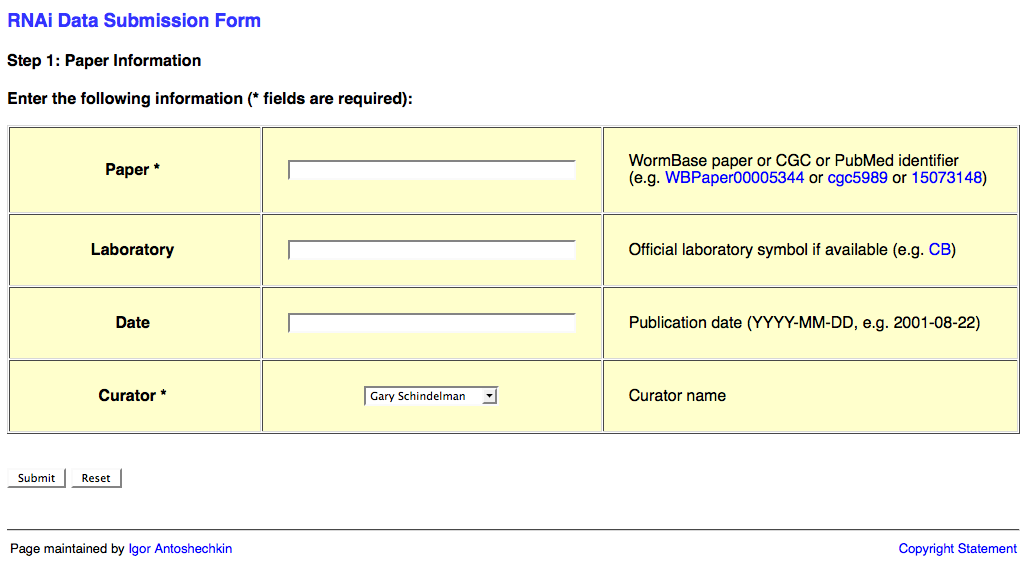
The first page of the web form looks like this:
This page requests information about the paper (e.g. WBPaperID), laboratory (usually senior/corresponding author; for a list of all labs two-letter codes look here: http://www.wormbase.org/db/misc/laboratory?name=*;class=laboratory), date of publication (accepted date), and a curator name.