Difference between revisions of "RNAi"
m (→(2) Batch Form) |
m (→(2) Batch Form) |
||
| Line 237: | Line 237: | ||
| − | The next 8 columns (G-N) record RNAi probe information (i.e. information about the dsRNA identity/sequence). The first column (G) is an exception. The "Same As Above" column either takes no entry or "YES" (all CAPS important); this column indicates whether the row/line of the form is to be included as part of the same RNAi experiment as the line(s) before it (the closest preceding line with paper information provided). It is important that information that is unique to an RNAi experiment (e.g. Strain/Genotype, Treatment, Temperature, etc.; indicated by RED text) is not also added to this line as it will overwrite any information provided in lines above it (belonging to the same RNAi experiment). | + | The next 8 columns (G-N) record RNAi probe information (i.e. information about the dsRNA identity/sequence). The first column (G) is an exception. The "Same As Above" column either takes no entry or "YES" (all CAPS important), available from an Excel pull down menu; this column indicates whether the row/line of the form is to be included as part of the same RNAi experiment as the line(s) before it (the closest preceding line with paper information provided). This is necessary, for example, when submitting more than one RNAi probe, as each RNAi probe must go on its own line. It is important that information that is unique to an RNAi experiment (e.g. Strain/Genotype, Treatment, Temperature, etc.; indicated by RED text) is not also added to this line as it will overwrite any information provided in lines above it (belonging to the same RNAi experiment). |
The "PCR Product", "Primer1", "Primer2", "Genomic Coordinates", "Clone", and "Sequence" columns record the dsRNA source as in the CGI Web Form (see above). The "Target ID" column (yellow, hence to be removed before submission) is free-text to indicate which gene the RNAi probe is targeting. This column is handy for keeping track of which RNAi experiments have already been curated, as there is no other easy way to see the gene's name in the form. | The "PCR Product", "Primer1", "Primer2", "Genomic Coordinates", "Clone", and "Sequence" columns record the dsRNA source as in the CGI Web Form (see above). The "Target ID" column (yellow, hence to be removed before submission) is free-text to indicate which gene the RNAi probe is targeting. This column is handy for keeping track of which RNAi experiments have already been curated, as there is no other easy way to see the gene's name in the form. | ||
| Line 246: | Line 246: | ||
[[File:RNAi_Batch_Form_3.png]] | [[File:RNAi_Batch_Form_3.png]] | ||
| − | + | The next 8 columns (O-V) record experimental details including "Strain", "Genotype", "Treatment", "Life Stage", "Temperature", "Delivered by", "Species", and "Remark". The "Life Stage" and "Delivered by" columns provide Excel pull down menus to select terms from a standard list (syntax is important). Note that these fields are the ones that are unique to an RNAi experiment; if two or more rows contain information for one of these columns, only the bottom-most row will be read. The formats for the remaining columns are as indicated in the CGI Web Form section above (i.e. "Genotype", "Treatment", and "Remark" are free-text, "Strain" must be a recognized WormBase strain name, "Temperature" must be an integer value for degrees Celsius, and "Species" must be a recognized Latin species name of a nematode species in WormBase). | |
Revision as of 22:15, 15 September 2011
Contents
RNAi Curation Mission Summary
The term "RNAi" stands for "RNA-interference" and refers to the targeted silencing of gene expression of a "target gene" via introduction of double stranded RNA (dsRNA) containing high degrees of sequence identity to the "target gene". In C. elegans, molecules of dsRNA may be introduced into the worm by a variety of different methods including direct micro-injection, soaking of worms in solution containing dsRNA, feeding worms bacteria that express dsRNA from a plasmid, and transgenic expression (within the worm) of dsRNA. For efficient and specific knockdown of gene expression, the dsRNA must have a minimum sequence identity with the target gene sequence. Any phenotype(s) resulting from the RNAi-mediated knockdown of a particular gene is thought to directly reflect the phenotype of a loss-of-function mutation in that gene, hence providing evidence as to the gene's biological function.
The goal of RNAi curation is to associate RNAi-mediated phenotypes with the target genes knocked down in RNAi experiments, as found in the literature pertaining to C. elegans and related species. There are some important things to consider when curating RNAi experiments. Some RNAi experiments are simple to curate as the strain is N2 (wild type genotype), only a single gene is targeted for RNAi, the phenotype is clearly stated, and the source and identity of the dsRNA is clearly stated by the authors. In many cases, however, the curation task is not so simple: there may be complex genotypes with multiple mutations in the strain receiving the dsRNA, multiple RNAi gene targets, complex genetic interactions, and/or missing descriptions from the authors as to controls, dsRNA delivery method, dsRNA identity, and/or phenotype directly resulting from the RNAi in question. Hopefully these issues will all be addressed below.
RNAi Data Model
This is the RNAi data model as of WormBase Release WS227:
//////////////////////////////////////////////////////////////////
//
// ?RNAi class
//
//////////////////////////////////////////////////////////////////
?RNAi Evidence #Evidence
History_name UNIQUE ?Text
Homol Homol_homol ?Homol_data XREF RNAi_homol ?Method Float Int UNIQUE Int Int UNIQUE Int #Homol_info
Sequence_info DNA_text Text UNIQUE Text //stores actual probe sequence for automated mapping
// 1st Text is DNA, 2nd is probe name
Sequence ?Sequence XREF RNAi //links to a real Sequence object used in the experiment
// such as yk clone; not UNIQUE anymore
PCR_product ?PCR_product XREF RNAi // links to a PCR_product object used in
// the experiment; not UNIQUE anymore
Uniquely_mapped //boolean; if present, signifies that ?RNAi object has a unique sequence
// which maps to a single place in the genome
Experiment Laboratory ?Laboratory
Author ?Author
Date UNIQUE DateType
Strain UNIQUE ?Strain
Genotype UNIQUE ?Text //used when no Strain object exists
Treatment UNIQUE ?Text
Life_stage UNIQUE ?Life_stage
Temperature UNIQUE Int
Delivered_by UNIQUE Bacterial_feeding //RL [010327]
Injection //RL [010327]
Soaking //RL [010327]
Transgene_expression //RL [010327]
Inhibits Predicted_gene ?CDS XREF RNAi_result #Evidence // "gene" parent (unreliable)
Gene ?Gene XREF RNAi_result #Evidence //RL [010327]
Transcript ?Transcript XREF RNAi_result #Evidence // [021126 krb]
Pseudogene ?Pseudogene XREF RNAi_result #Evidence // [030801 krb]
Supporting_data Movie ?Movie XREF RNAi // Lincoln, krb [010807]
DB_info Database ?Database ?Database_field ?Accession_number //to link out to Phenobank ar2 02-DEC-05
//removed UNIQUE as reqs multiple connections
Species UNIQUE ?Species
Gene_regulation ?Gene_regulation XREF RNAi // this tag is used when an RNAi experiment describes
// gene regulation ar2 29-MAR-06 for igor
Interaction ?Interaction
Reference UNIQUE ?Paper XREF RNAi //[070215 ar2] made reference unique so Paper sort of
// equates to a Study class for Will S
Phenotype ?Phenotype XREF RNAi #Phenotype_info
Phenotype_not_observed ?Phenotype XREF Not_in_RNAi #Phenotype_info //added by Wen to separate
// Not phenotype from real phenotypes
Expr_profile ?Expr_profile XREF RNAi_result // connection added during build [030106 krb]
Remark ?Text #Evidence
Method UNIQUE ?Method
RNAi Curation Standard Operating Procedure (SOP)
In order to ensure consistency of RNAi curation across curators and WormBase releases, a set of standard procedures for RNAi curation are outlined below. The descriptions of these procedures include use of the two main methods for generating *.ace files for release submission: (1) the web-based CGI form for one-at-a-time RNAi object generation and (2) the batch form submission method.
Minimum Requirements for an RNAi Object
Regardless of which curation method you (the curator) choose, there are a set of minimum requirements in order to generate a complete RNAi experiment object:
1) A reference (e.g. WBPaperID)
2) A curator name
3) The sequence of the dsRNA used in the experiment to knockdown gene expression
4) A dsRNA delivery method (e.g. Injection)
5) A phenotype (observed or not_observed)
If any one of these five basic pieces of information are missing, the scripts that generate the *.ace file will fail, returning an error.
Two Methods of Curation: Web CGI & Batch Form
There are two basic methods of RNAi curation: (1) a web-based CGI form and (2) a batch form involving a Perl script (living on the elbrus@caltech machine) to convert a spreadsheet (tab-delimited file) into an *.ace file.
(1) Web-based CGI
The web-based RNAi curation form can be found at:
http://elbrus.caltech.edu/cgi-bin/igor/rnaitools/rnai_curation
Web Form Page One
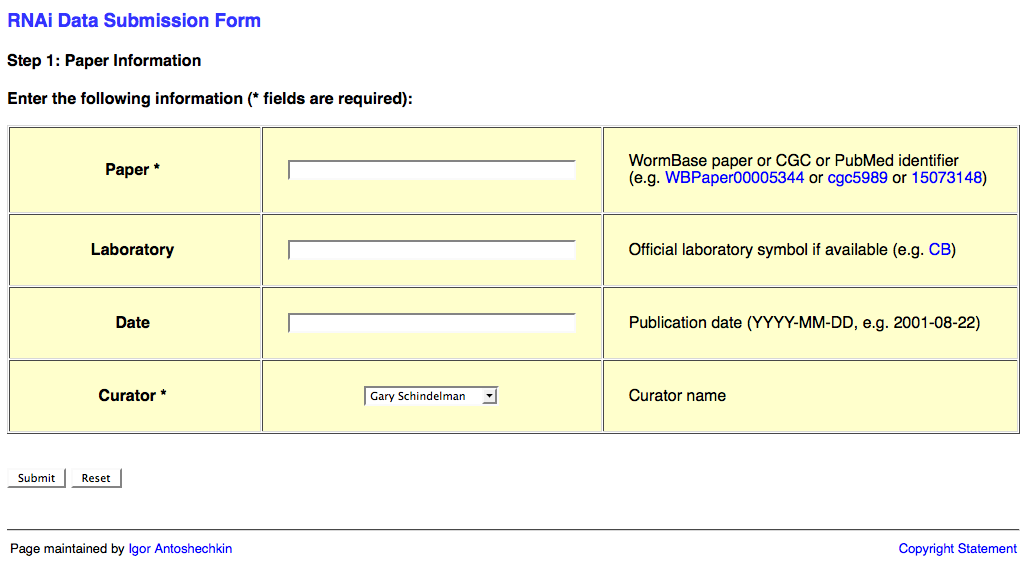
The first page of the web form requests information about the publication and looks like this:
This page requests information about the paper (e.g. WBPaperID), laboratory two-letter code (usually senior/corresponding author; for a list of all labs' two-letter codes look here: http://www.wormbase.org/db/misc/laboratory?name=*;class=laboratory), date of publication (accepted date), and a curator name. The laboratory and publication acceptance date are optional.
Once you've input the relevant information, click on the "Submit" button. Note that this may take a few moments to load. Once loaded you will be at the second page of the web form.
Web Form Page Two
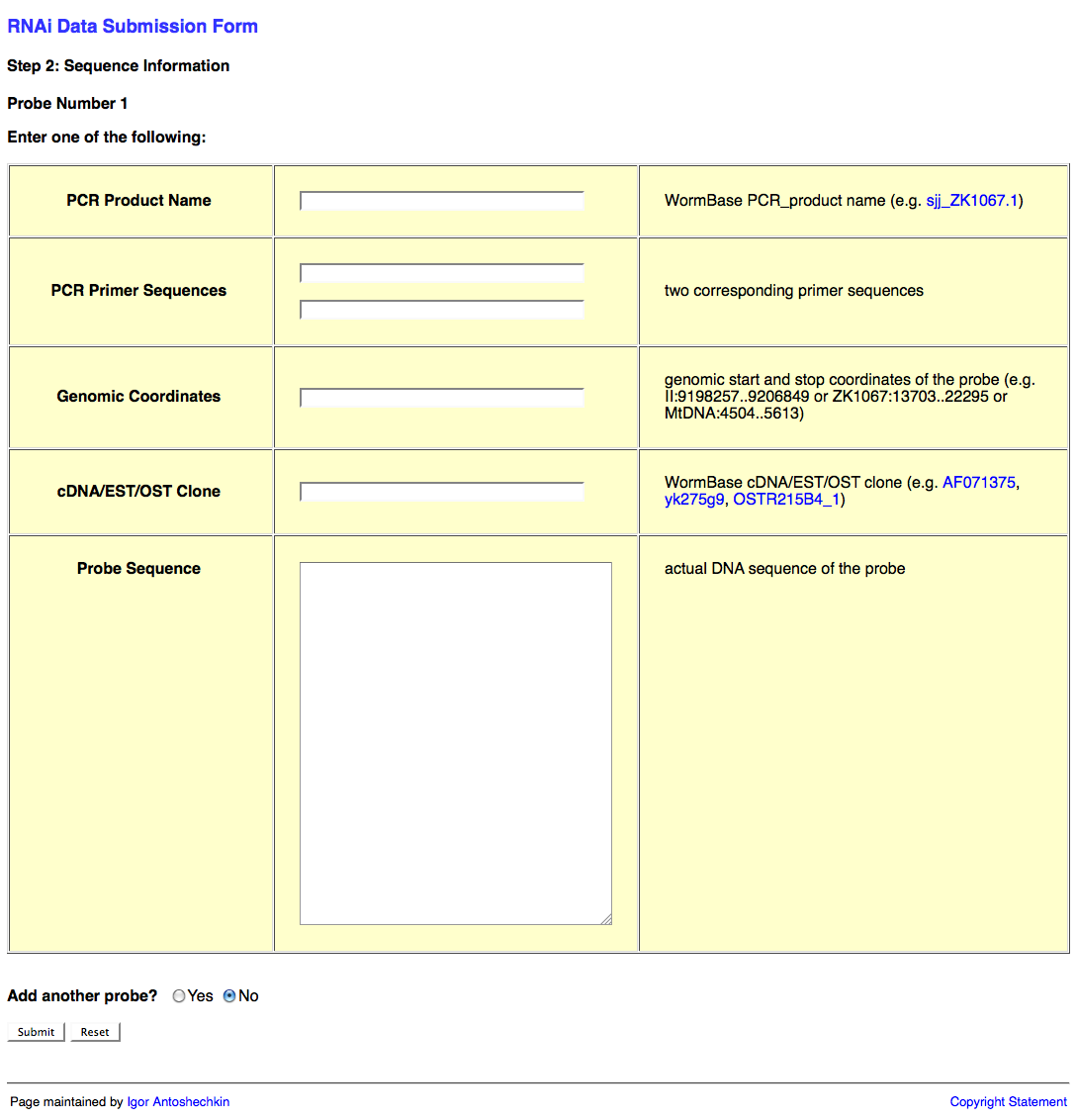
The second page of the web form looks like this:
This second page of the web form requests information about the RNAi probe(s) (i.e. the dsRNA sequence used to knockdown expression of the target gene). This information may be submitted in the form of a PCR product object (e.g. sjj_ZK617.1 or mv_CAA33463), PCR primers (it's usually good practice to check the primer sequences using e-PCR to confirm that they work properly), genomic coordinates, cDNA/EST/OST clone (note that many of these clones may be problematic; in such a case it is best to determine the sequence to the best of your ability and submit that instead), or actual sequence (e.g. "ACGT") of the probe.
An important point here is that RNAi target genes for an RNAi experiment are not submitted directly using the name of the target gene as indicated in the publication. The reason for this is that gene names can be continuously remapped to different regions of the genome over time, making persistence of the data impossible if we were to use only gene names to identify the target gene. By providing the actual sequence or clone used in the experiment, we can ensure that the appropriate gene is identified as the RNAi target gene regardless of which release of the database you are currently working in.
You may wish to submit more than one RNAi probe (e.g. when two or more genes are targeted by RNAi simultaneously). If this is the case, be sure to select "Yes" after the "Add another probe?" question at the bottom of the page, and then click on the "Submit" button.
Once all of your probes have been submitted, make sure that "No" is selected after the "Add another probe?" question, and click on the "Submit" button.
If all of the probes check out OK, you will be brought to the third page of the web form.
Web Form Page Three
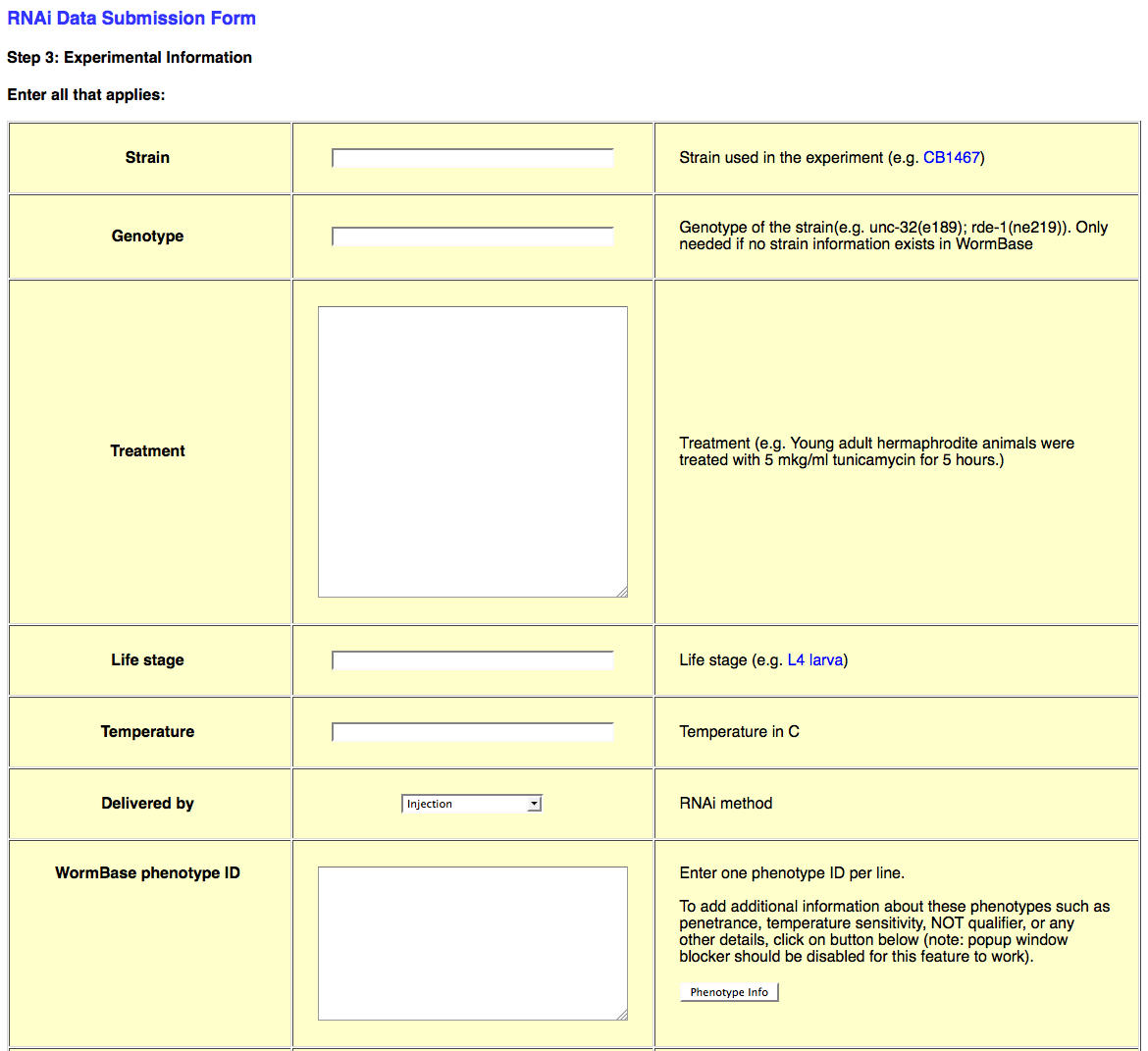
The third page of the web form looks like this:
This page requests all of the experimental details about the RNAi experiment in question. This information includes Strain/Genotype, treatment conditions, life stage, dsRNA delivery method (e.g. bacterial feeding), Phenotype observed (or not_observed), Gene Regulation objects, Genetic Interaction objects, species, and general remarks about the experiment.
Strain/Genotype
Whereas the "Genotype" information for a strain used in an RNAi experiment may be written in as free-text, the "Strain" field will only accept strains that are officially recognized by WormBase. Be sure to check the database for the existence of your strain name before submitting (otherwise you may receive an error upon submission). If your strain name does not currently exist in the database, you may write in the genotype in the "Genotype" field, or request the addition of the strain into the database.
Treatment/Temperature
The "Treatment" field is a free-text field in which the curator may specify any particular experimental conditions that apply to this RNAi experiment. This may include details about growth conditions, dsRNA delivery, or specimen manipulation. The "Temperature" field must be filled in with an integer value indicating the temperature in degrees Celsius (e.g. "25" for 25 degrees Celsius).
Life Stage
The "Life Stage" field must be filled in with an official life stage name, as stored in ACeDB/WormBase. For an official list of Life Stage names in WormBase, see:
or
Delivered by
The "Delivered by" field provides a drop-down menu of four dsRNA delivery methods: Injection, Bacterial feeding, Soaking, and Transgene expression.
WormBase phenotype ID
This field needs to have at least one phenotype in the form of a WormBase phenotype ID, for example: WBPhenotype:0000050 (for embryonic lethality). Without at least one phenotype, the *.ace-generating script will throw an error.
Additional phenotype information...
If you cannot find an appropriate phenotype for your RNAi experiment in the Worm Phenotype Ontology (WPO; worm_phenotype.obo can be downloaded from http://www.obofoundry.org/), you may provide a description of the phenotype here so that a new phenotype will be generated within the WPO. Be sure to check the "New Phenotype" box.
This field may also be used to describe additional details about the phenotype that may not be evident from the WBPhenotype ID chosen for curation.
New Gene Regulation
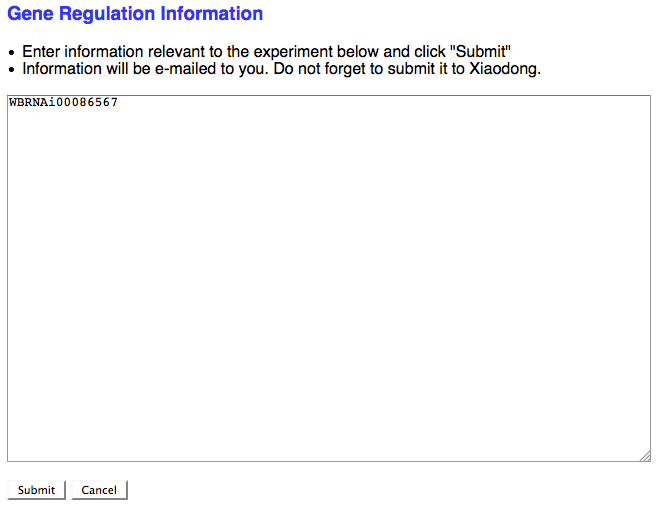
If the RNAi experimental result suggests that the RNAi target gene is somehow functionally involved in the regulation of a gene (at the transcriptional, translational, or post-translational level) and no Gene Regulation object exists for this relationship in the database, check the box here to request that a Gene Regulation object ID be assigned to this RNAi experiment. If the curator checks the box, a separate browser window will open with a simple text field to enter in the details of the Gene Regulation event:
As the note suggests, don't forget to send this info to Xiaodong so she can generate a new Gene Regulation Object.
Gene Regulation Object
If a Gene Regulation Object ID has already been assigned to this experiment from a different curation pipeline, enter the Gene Regulation Object ID here (e.g. WBPaper00006370_lin-3, cgc6303_lag-2).
New Interaction
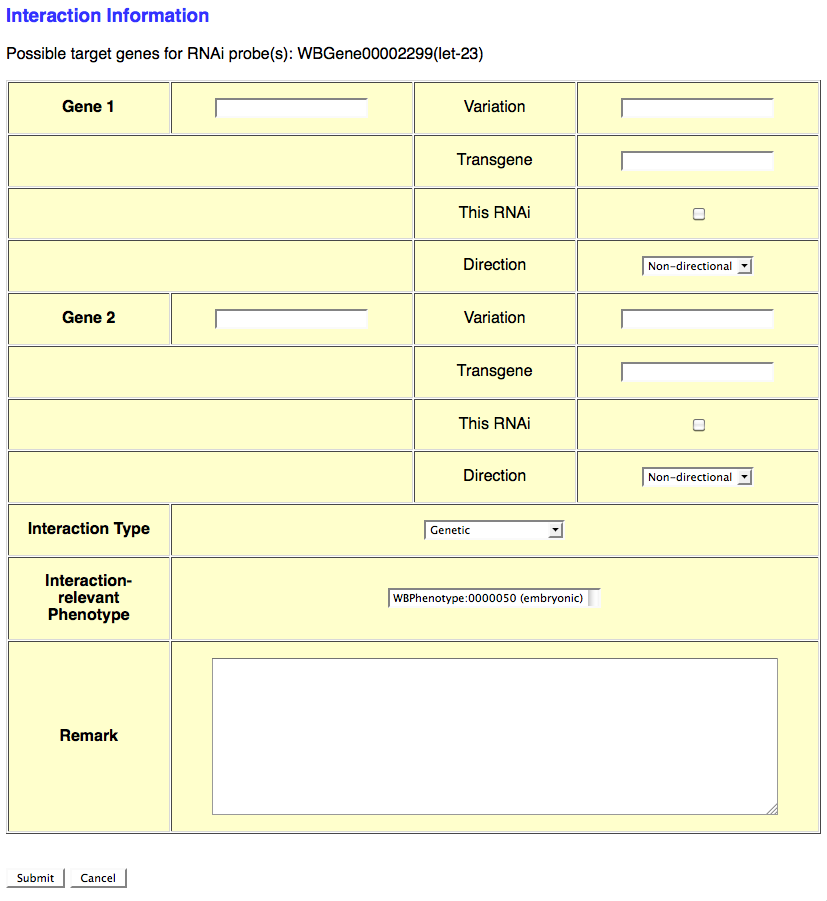
If the RNAi experiment indicates a genetic (or any other type) interaction and an Interaction Object ID has not yet been assigned to this interaction, select the number of genes involved in the interaction from the provided pull down menu. This will automatically open another browser window which will provide a form for entering details specific to that particular interaction (see image below):
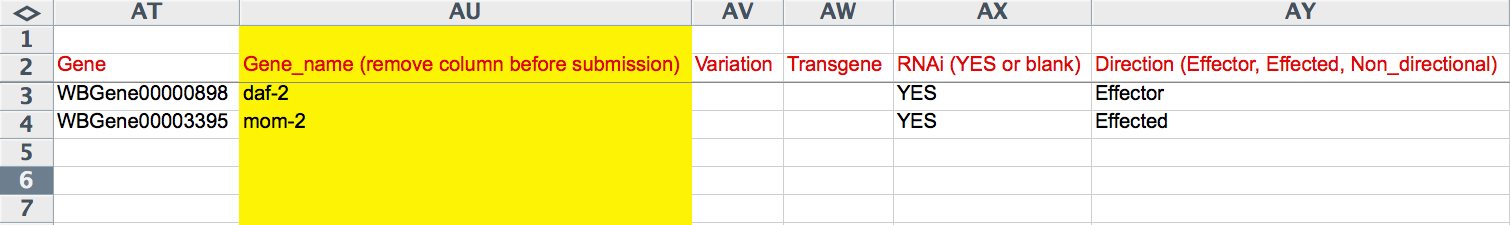
This form provides the curator with fields to enter information for each interacting gene as well as general information about the interaction. The gene-specific information includes the gene's WBGeneID (e.g. WBGene00002299), "Variation" information (e.g. e1370, mgDf47) if this gene was perturbed by an allele or other variation, "Transgene" for a transgene harboring the gene (or variation of the gene) that contributes to the interaction, "This RNAi" toggle box to indicate that the perturbation of this gene was via RNAi (as opposed to an allele), and "Direction" drop-down menu to indicate if the gene is the "Effector" or "Effected" gene (for directional interactions) or "Non-directional" (for interactions in which directionality is irrelevant).
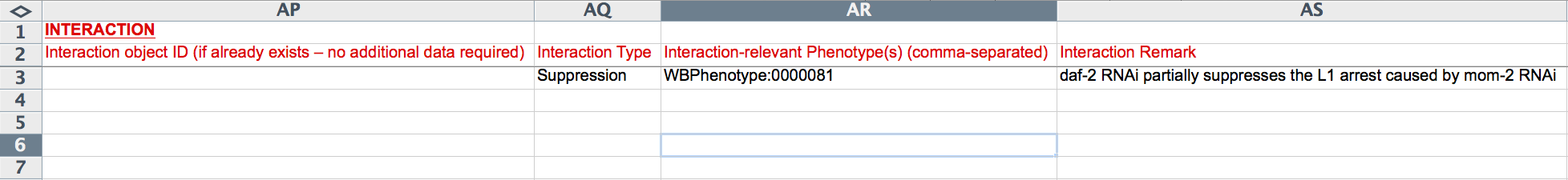
Below the gene-specific fields is a drop-down menu for indicating the "Interaction Type" (e.g. Genetic, Suppression, Enhancement), a "Interaction-relevant Phenotype" field to indicate the phenotype about which the interaction is based, and a free-text "Remark" field to describe any details of the interaction not sufficiently explained by the other fields.
Interaction Object
If an Interaction Object ID has already been assigned to this experiment from a different curation pipeline, enter the Interaction Object ID here (e.g. WBInteraction0001295).
Picture
OBSOLETE; The current RNAi data model no longer supports Picture objects
Species
Enter the species in which the RNAi experiment is taking place. Caenorhabditis elegans is set by default.
Remark
The "Remark" field is a free-text field to enter in any remaining relevant information that is pertinent to the RNAi experiment, but has not yet been captured by any other field of the curation form. This is often where standard remarks regarding the ambiguity of the dsRNA identity (see below) are entered. Information relevant to a genetic interaction, gene regulation event, or complex phenotype may also be put here.
If you are done curating this particular RNAi experiment and have more from the current paper, select "Yes" next to the "Add another experiment from this paper?" question at the bottom of the page and then click "Submit". If you are done curating RNAi experiments from this paper (or are done for the current session) and would like to generate an *.ace file, select "No" next to "Add another experiment from this paper?". Finally, indicate whether or not you would like to receive an e-mail of the *.ace file (as text in the body of the e-mail) next to "Would you like to receive a copy of the ace file via e-mail?". Once you are set, click the "Submit" button to generate the ace file (to be displayed in the web browser). To generate the *.ace file, you may simply copy and paste the text from the screen into a plain text file and save it with the ".ace" extension.
(2) Batch Form
The second basic method for generating RNAi *.ace files involves filling out all relevant RNAi curation information on a spreadsheet with each row (more or less; explanation below) as an RNAi experiment and each column as an RNAi object field. An Excel-based template file may be found here:
File:RNAi Curation Template.xls
Much of what applies to the RNAi Curation CGI Form described above applies similarly (or exactly) to the Batch Form. There are a few spreadsheet-specific changes that we will describe here.
First, there are a number of columns that are highlighted in yellow and have the comment "remove column before submission" in parentheses. These are temporary aids to the curator that must be removed before submitting the spreadsheet to the script, as they will cause an error.
Shown below are screenshots from a fictional RNAi experiment entry in the batch form.
Paper Identification Information
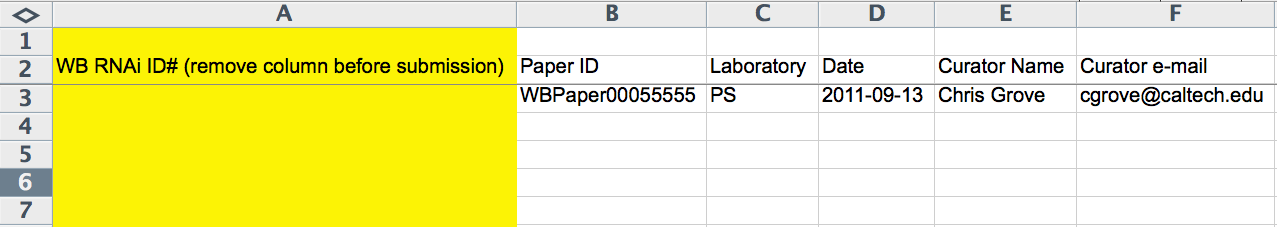
The first 6 columns (A-F) of the spreadsheet record WBRNAi ID, WBPaper ID (WBPaper000#####), Laboratory (2-letter code), Publication Acceptance Date (YYYY-MM-DD), Curator Name, and Curator E-Mail. The WBRNAi ID column is only used once the *.ace file has already been made and is removed before submission to the *.ace-file-generating script. Whereas the Laboratory code and Publication Acceptance Date are optional, the WBPaper ID and the Curator Name are required.
RNAi probe (dsRNA source) Information
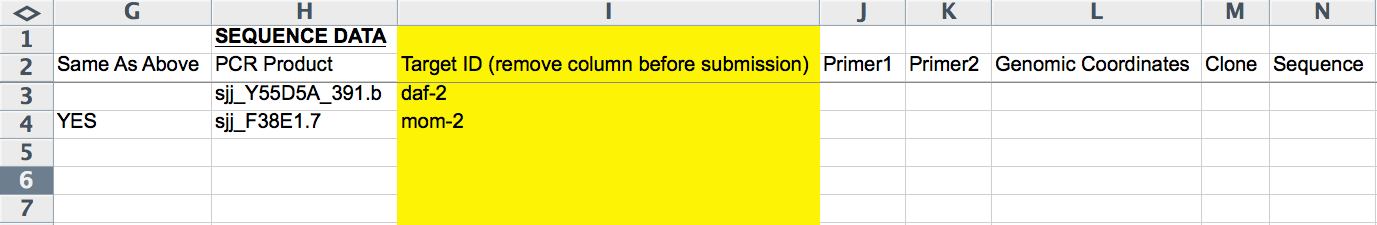
The next 8 columns (G-N) record RNAi probe information (i.e. information about the dsRNA identity/sequence). The first column (G) is an exception. The "Same As Above" column either takes no entry or "YES" (all CAPS important), available from an Excel pull down menu; this column indicates whether the row/line of the form is to be included as part of the same RNAi experiment as the line(s) before it (the closest preceding line with paper information provided). This is necessary, for example, when submitting more than one RNAi probe, as each RNAi probe must go on its own line. It is important that information that is unique to an RNAi experiment (e.g. Strain/Genotype, Treatment, Temperature, etc.; indicated by RED text) is not also added to this line as it will overwrite any information provided in lines above it (belonging to the same RNAi experiment).
The "PCR Product", "Primer1", "Primer2", "Genomic Coordinates", "Clone", and "Sequence" columns record the dsRNA source as in the CGI Web Form (see above). The "Target ID" column (yellow, hence to be removed before submission) is free-text to indicate which gene the RNAi probe is targeting. This column is handy for keeping track of which RNAi experiments have already been curated, as there is no other easy way to see the gene's name in the form.
Experimental Information
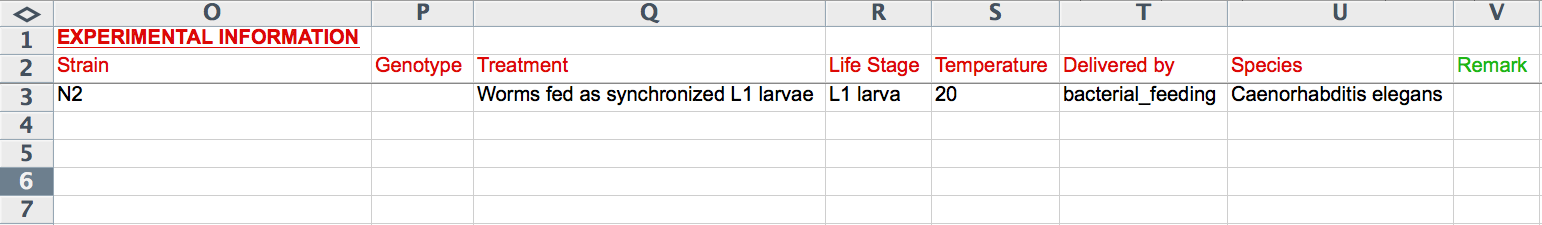
The next 8 columns (O-V) record experimental details including "Strain", "Genotype", "Treatment", "Life Stage", "Temperature", "Delivered by", "Species", and "Remark". The "Life Stage" and "Delivered by" columns provide Excel pull down menus to select terms from a standard list (syntax is important). Note that these fields are the ones that are unique to an RNAi experiment; if two or more rows contain information for one of these columns, only the bottom-most row will be read. The formats for the remaining columns are as indicated in the CGI Web Form section above (i.e. "Genotype", "Treatment", and "Remark" are free-text, "Strain" must be a recognized WormBase strain name, "Temperature" must be an integer value for degrees Celsius, and "Species" must be a recognized Latin species name of a nematode species in WormBase).
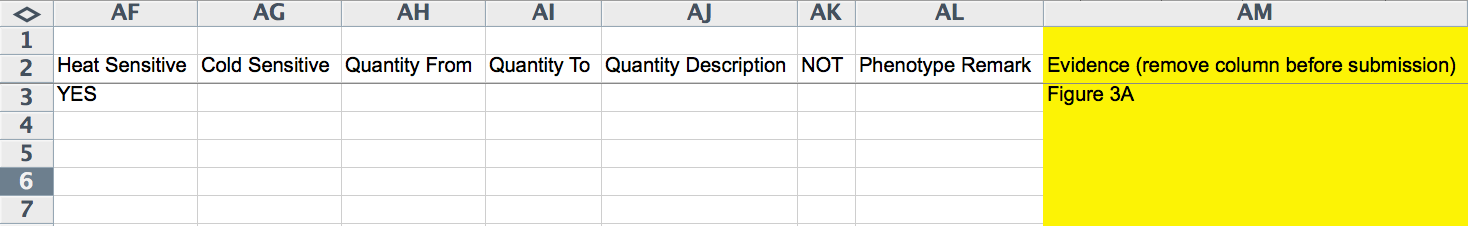
Phenotype Information 1
Phenotype Information 2
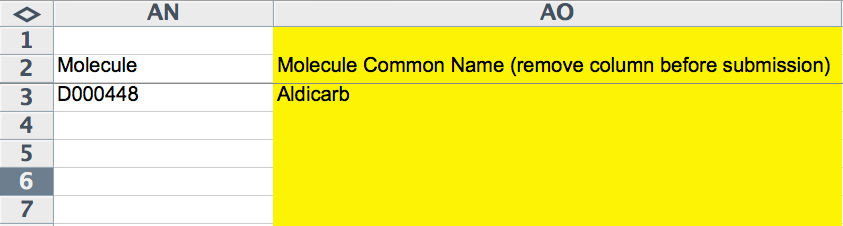
Molecule Information
Interaction Information 1
Interaction Information 2