Search results
From WormBaseWiki
Jump to navigationJump to searchCreate the page "Other species" on this wiki! See also the search results found.
Page title matches

File:Phenotype form RNAi field and nested species field 1-20-2016.png Screenshot of the RNAi and nested species fields in the Phenotype Form(642 × 115 (26 KB)) - 23:15, 20 January 2016
Page text matches

File:Wb species tree2.png updated species tree(309 × 239 (6 KB)) - 13:20, 24 September 2010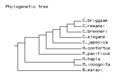
File:Wb species tree.png WormBase species as of WS219(287 × 191 (5 KB)) - 13:06, 24 September 2010- ?Species Common_name ?Text Protein ?Protein XREF Species411 bytes (44 words) - 09:14, 17 August 2010
- ===Nematode Species Mentioned in Titles and Abstracts of Current Papers===525 bytes (60 words) - 15:34, 17 May 2016

File:Treeprint2.png Phylogenetic Tree of nematode species in WormBase(518 × 435 (10 KB)) - 08:58, 9 June 2011
File:Treeprint3.png phylogenetic tree of the WormBase species. Made using Phylodendron.(508 × 424 (4 KB)) - 10:37, 8 December 2011- *Survey organism databases for methods of presenting and navigating between species. *Codify best practices for presentation of multiple species data.885 bytes (118 words) - 16:42, 9 November 2010
File:Phenotype form RNAi reagent and species fields Feb 2018.png Screenshot of the phenotype form RNAi reagent and species fields(814 × 176 (34 KB)) - 23:01, 13 February 2018- === Support for 'non-canonical' species at the Alliance === ...ical issues surrounding support for 'non-canonical' species (those species other than the main 8 currently supported at the Alliance)2 KB (348 words) - 17:52, 2 February 2023
- ===Handling species lists=== * Handling species in WormBase and Alliance3 KB (440 words) - 08:42, 3 September 2020

File:Phenotype form RNAi field and nested species field 1-20-2016.png Screenshot of the RNAi and nested species fields in the Phenotype Form(642 × 115 (26 KB)) - 23:15, 20 January 2016- ||/Gene/Identity/Species/Species962 bytes (119 words) - 20:15, 2 May 2013
- ...nding page -e.g. under species menu. Show both assemblies. Advertise other species genomes on blog/news. * Discussion on giving WormBase IDs for other species. Michael P: for people tend to use genebank IDs is not obvious to map/searc929 bytes (147 words) - 19:14, 21 February 2013
- ...ember of the Elegans group, but the Elegans group's closest known outgroup species. It was selected for genomic sequencing on account of its providing the bes 1. A [http://dx.doi.org/10.1163/156854102321122557 species description article] in [http://www.ingentaconnect.com/content/brill/nemy '2 KB (264 words) - 16:01, 15 December 2011
- Origin Species UNIQUE ?Species930 bytes (84 words) - 14:02, 4 October 2010
- ...o species objects as the taxon I'd in the file is different to that in the species object in WormBase.704 bytes (104 words) - 12:27, 28 January 2014
- = Species Collection = It represents the species list as returned by the API.998 bytes (168 words) - 15:29, 19 June 2014
- == 3. Cross species searching; debug GBrowse search (Norie; 10 minutes) == http://dev.wormbase.org:9004/species/gene/WBGene00006763681 bytes (71 words) - 21:26, 12 January 2011
- ''Caenorhabditis brenneri'' is a gonochoristic (male-female) species closely related to ''C. elegans'' (i.e., a fellow member of the Elegans gro ...t has had several formal or informal synonyms, such as "''Caenorhabditis'' species 4" ("''C''. sp. 4"), "''C''. sp. CB5161" (or just "CB5161"), and "''C''. sp2 KB (291 words) - 15:59, 15 December 2011
- ''Caenorhabditis remanei'' is a gonochoristic (male-female) species closely related to ''C. elegans'' (i.e., a fellow member of the Elegans gro Some earlier studies of non-''elegans'' species occasionally misidentified ''C. remanei'' as "''C. vulgaris''", or confused2 KB (229 words) - 15:56, 15 December 2011