UserGuide:WormMine
WormMine is an integrated search tool of WormBase data built with the Intermine data warehouse platform.
(open a parallel browser window and open WormMine)
Search: type in an identifier or keyword and GO.
Templates: 'canned' queries that others have found useful. The easiest way to learn WormMine query is to modify and customize existing templates. You can create and save your own templates after logging in.
Lists: operate on whole lists of data at once. You can upload lists or save them from results tables. Useful public lists are provided.
QueryBuilder: create custom queries and edit existing templates. QuiryBuilder is very powerful but may demand some learning.
MyMine: create an account to save your own queries, lists and templates, as well as marking public templates as favorites.
Contents
Template Query
WormMine provides a library of public template queries - predefined queries designed to perform a particular task. Each one has a description and a form to fill in. For example, there are templates to find genes annotated with a specific GO (GeneOntology) term, genes expressed in a specific tissue (anatomy term).
You can find all WormMine templates on the templates page, accessible via the toolbar on any page. Here you can search the titles and descriptions of all templates.
Anyone can add private (for-your-eyes-only) templates. To create a template:
- Log in
- Construct a query using Query Builder
- Must include at least one constrain condition (e.g. restrict the format of the identifier)
- "Start building a template query"
- Fill in Name, Title and Description, and optionally comment
- Make necessary adjustments, then "Save template"
Lists
WormMine can operate on custom lists of data. You can save lists from results pages or create them by uploading lists of identifiers. Lists can be used when running template queries and analyzed by a series of widgets on a list analysis page. You can merge, subtract and find common members if you have more than one list.
WormMine includes a set of 'Public' lists (marked with 'PL') that we think might be useful for our users. Some of these originate from external data sources.
All lists, public ones as well as personal ones (if you are logged in) can be viewed on the Lists page, where you can search them and do operations on them.
To create a new list yourself, click on 'Lists', and then on 'Upload' in the toolbar on any WormMine page:
WormMine's list creation tool helps you upload a list of identifiers, the list can contain a mix of identifier types.
To preserve a list from query:
- Log in.
- Make a query (Query Builder).
- On the Result page, top right corner, select "Create / Add to List" -> "Create New List" -> "All of Columns ...", or "Choose individual items from the table" for further refinement.
- Provide for the list: a Name, an informative Description, and Tags. Add one tag at a time. Tags help to group lists. Hit "Create".
- Descriptions and tags can also be edited after a list is saved.
QueryBuilder
Use case: How do I find all proteins for a gene?
How are they intuitively connected? Genes are transcribed into transcripts which contain coding sequences which are translated into proteins. Keep this in mind while constructing a query.
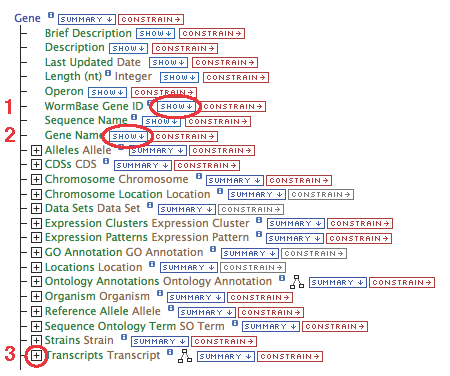
- Select "Gene" in the box under "Select a Data Type to Begin a Query"
- Click "Select", or double click "Gene"
This list represents the Gene data available to display. Various fields can be chosen to be shown in the results table, links to other data types followed, and filters can be set to constrain the results.
Desired results?
We want to display the symbols of each involved object, plus the gene ID. This calls for:
- Gene.WormBase Gene ID
- Gene.Gene Name
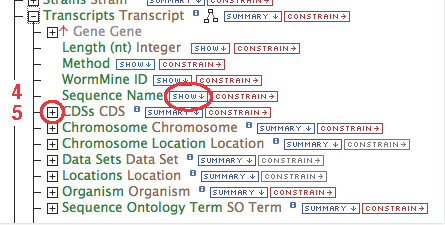
- Gene (some relationship(s))-> Transcript.Sequence Name
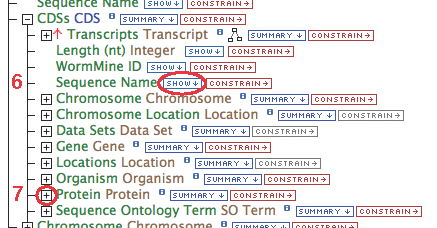
- Gene (some relationship(s))-> CDS.Sequence Name
- Gene (some relationship(s))-> Protein.Name
Building the query
Click *SHOW* next to any of these field names to add that attribute to the resulting table as a column.
- Show the "WormBase Gene ID" and "Gene Name"
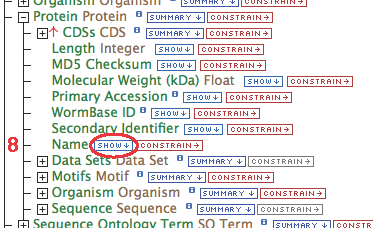
- Gene contains a collection of transcripts, this relationship represents transcription. Expand the relationship by clicking on the
[+]
- Follow these steps for each of the data types in the chain as illustrated:
- Running the query at this point will give you results for all genes. If you have a smaller set in mind, like restricting the genes to egl-19 only, a constraint must be set.
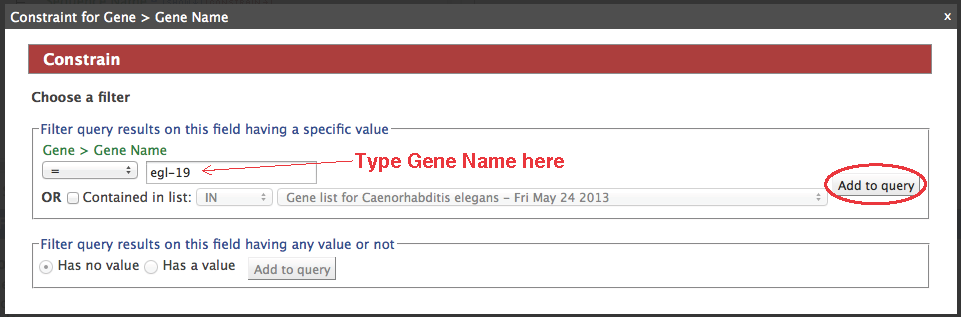
- Click "constrain" next to the "Gene.Gene Name" field (circle #2), type "egl-19" in the text box, make sure the operator is "=", then "add to query".
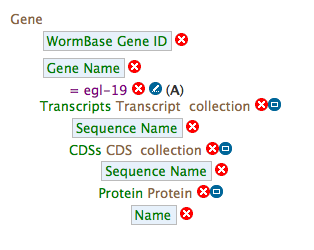
- Your query overview should resemble this:
- Show results will execute the completed query.
constraining to a list
If you have access to any lists, the constraint dialog box will provide options with respect to them. Fields can be constrained such that their values must or mustn't be a member of that list.
MyMine
All lists and queries you ran will be saved temporarily in WormMine for the current session. To save them permanently, you can create a MyMine account. You only need to provide an email address and a password to generate an account, there is no other information required. Your saved data is always private.
You can then access all your lists, queries and templates via the MyMine page. In MyMine you can save lists and queries you create in the QueryBuilder. You can even use the QueryBuilder to turn queries into new templates of your own. You can export/import queries and templates as XML to share them with others.