Lists
WormMine Lists
Lists in WormMine are a series of multiple items that can be either created by the user or provided by WormMine. Lists can be used as query constraints in new queries or templates, and they can be used to calculate unions, intersections and subtractions with other lists. WormMine lists can accept most types of IDs present in the database, and they can be created from multiple types of IDs at the same time (i.e. Uniprot name and WormBase protein IDs). In order to take full advantage of lists, it is recommended that the user is logged in to their account.
Creating Lists
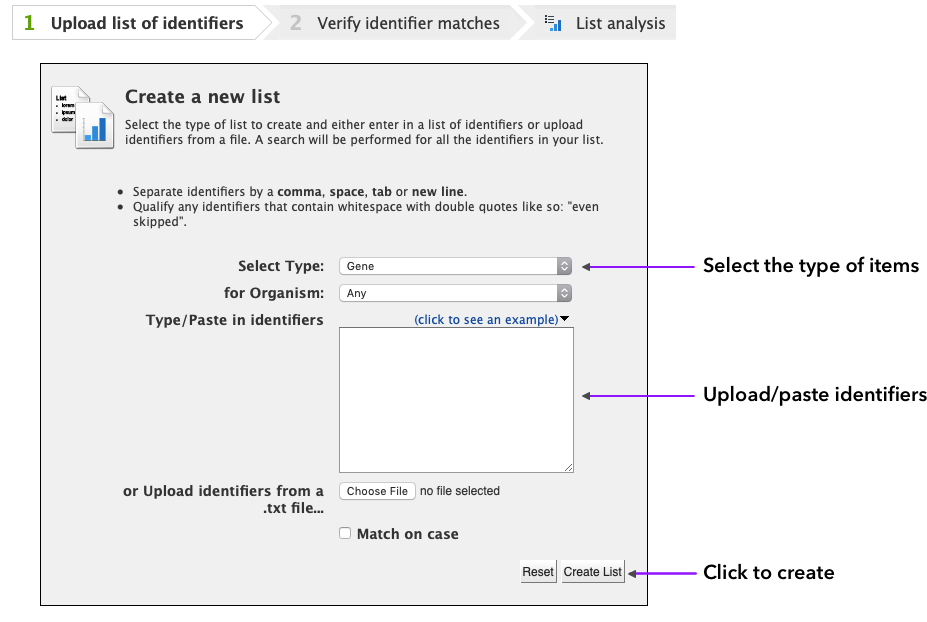
To create a list, click on the Lists tab on the main page. This will bring up the first page of the List creation wizard in WormMine.
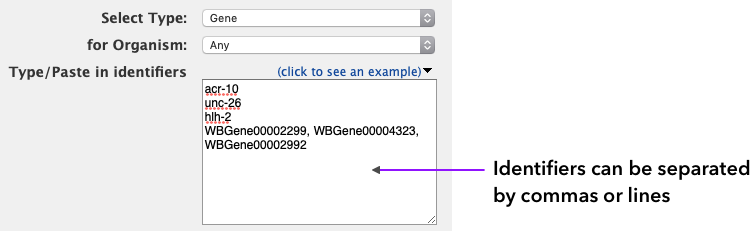
You can select the list type (from all the types of items in the database) and organism (majority of items are C. elegans). Enter or paste the identifiers (alternatively, a file can be uploaded) separated by comma, space, tab or new line and click on Create List.
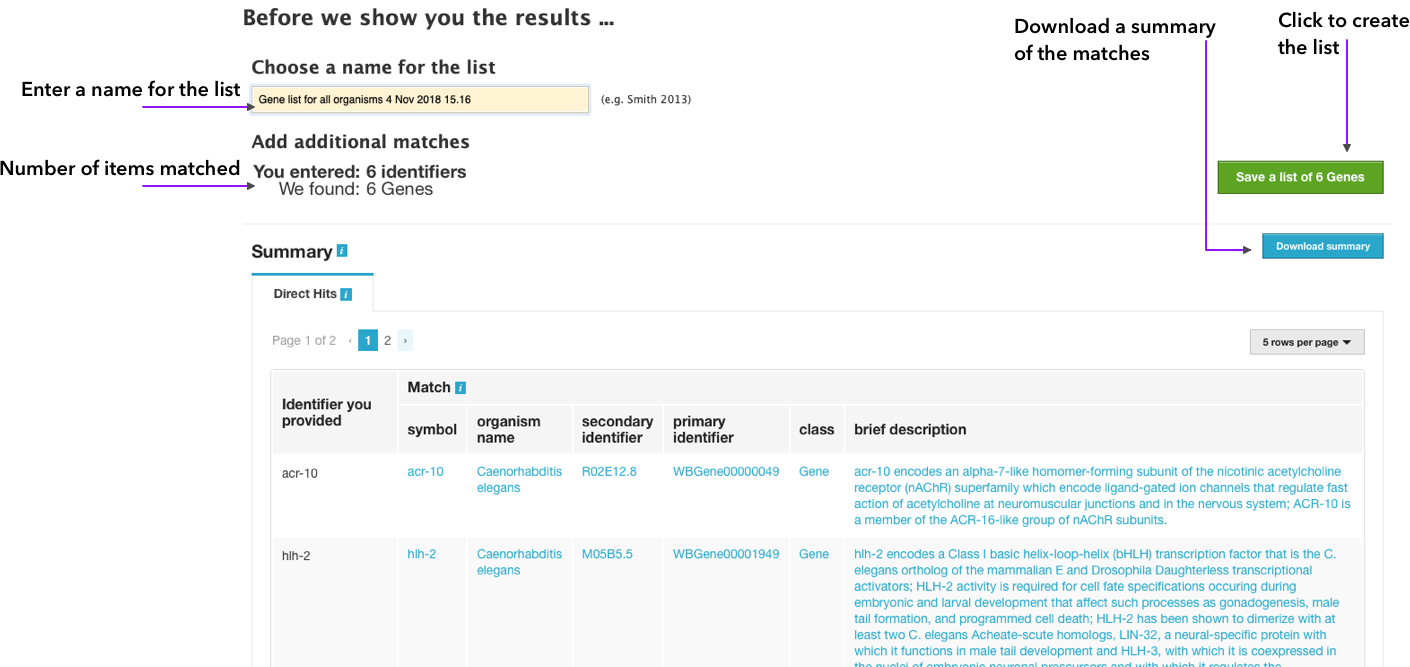
All the identifiers entered will be matched against the database and the second page of the wizard will be displayed.
A list name can then be entered, and the list created by clicking on Save a list of # items. There's a summary of the matches found, and you can download a CSV file with it. By saving the list, the final page of the wizard is shown.
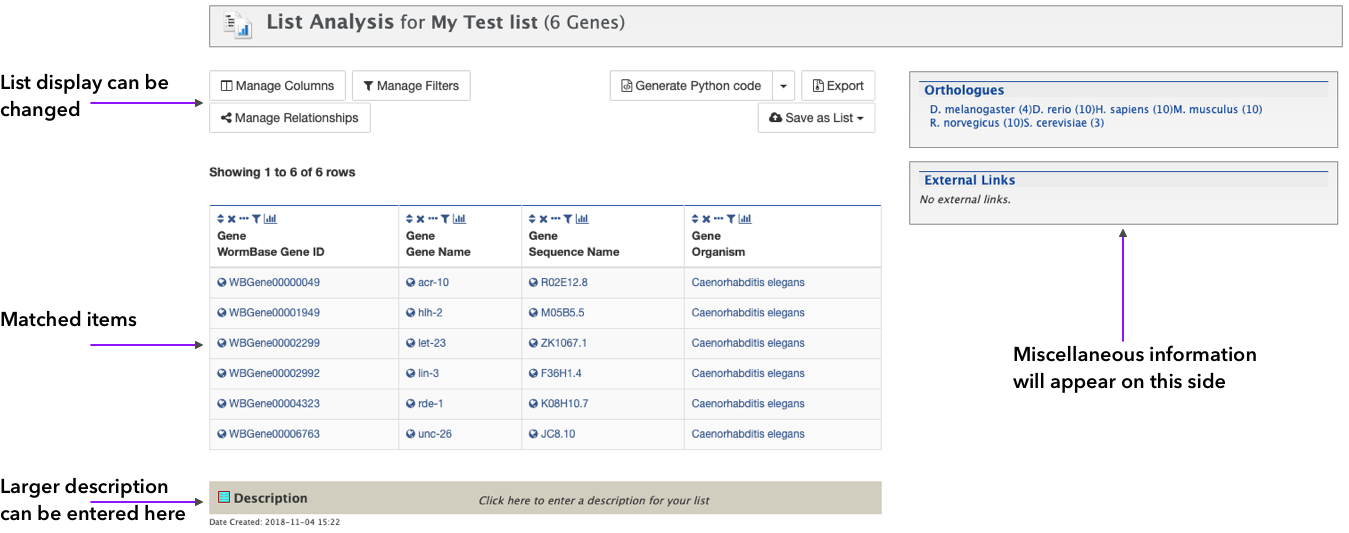
This page has similar functionalities as the query result table: the final list can be manipulated or changed, and other information can be entered, like a description for documentation, among other things. If you are logged in, the list will show in your My Mine lists where it can be retrieved anytime.
Using a list
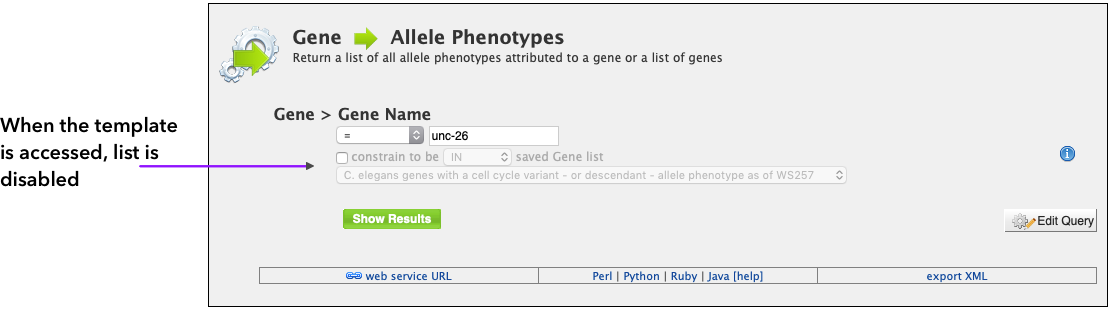
Lists can be used as constraints in different queries, as long as there is a correlation between the query and the type of items that compose a list. One example is applying a public list available in WormMine to the Gene --> Allele Phenotypes template query
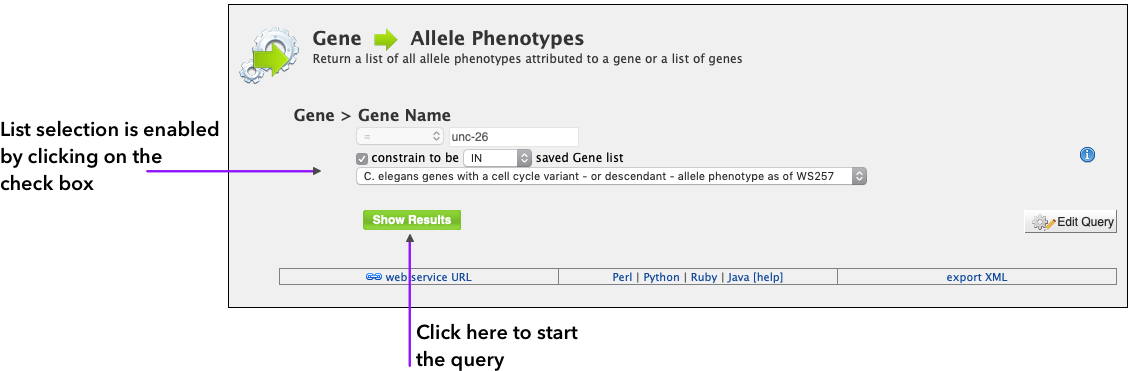
When the template is initially accessed, the list constraint option is disabled, allowing the user to choose a Gene Name to search. By clicking on the constraint to be the list dropdown is enabled, and the user can select a list to work as a constraint. By using the list in the query, the user has the option to search for a larger number of genes at the same time, and is not constrained by only one gene name as the normal template allows.