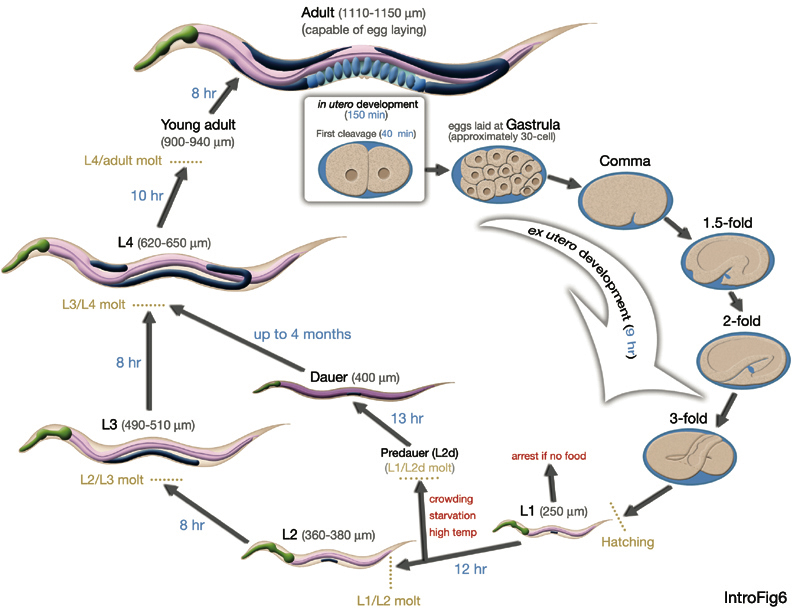
Life Stage
Contents
Life_Stage Data Model
////////////////////////////////////////////////////////////////////////////////////
?Life_stage Public_name UNIQUE Text //added by Daniela 120509
Contains Cell ?Cell XREF Life_stage
Cell_group ?Cell_group XREF Life_stage
Anatomy_term ?Anatomy_term XREF Life_stage
Sub_stage ?Life_stage XREF Contained_in
Contained_in ?Life_stage XREF Sub_stage // changed for Wen 010516
Preceded_by ?Life_stage XREF Followed_by
Followed_by ?Life_stage XREF Preceded_by
Expr_pattern ?Expr_pattern XREF Life_stage
Remark ?Text #Evidence
Definition UNIQUE ?Text #Evidence// added for Wen [020607 krb]
Other_name ?Text // added for Wen [020607 krb]
Reference ?Paper XREF Life_stage
Curated_by UNIQUE Text // added for Sylvia [010928 krb]
///////////////////////////////////////////////////////////////////////////////////
obo file
https://raw.github.com/draciti/Life-stage-obo/master/worm_development.obo
Life_Stage
Originally Life_stage objects were stored as names. In May 2012 Caltech decided to switch Life_stage objects from names to IDs. In order to do that we need a model change to store the original name (see model).
All data types that were using Life_stage had to be modified and dumped into Ids instead of names
- Phenotype -Karen
- Process -Karen
- RNAi -Chris
- gene regulation -Xiaodong
- gene expression -Daniela
- Anatomy_term -Raymond
- Expression Cluster -Wen
- Condition -Wen & Gary W
- Strain -Mary Ann
Life_Stage tags
tags used in Life_stage extracted from the Life_Stage. ace dump May 9th 2012
- Definition
- Anatomy_term
- Reference
- Other_name
- Expr_pattern
- Contained_in -> Change data into IDs
- Cell_group
- Sub_stage -> Change data into IDs
- Cell
- Followed_by -> Change data into IDs
- Remark -> goid: WBls could be deleted because will be redundant with IDs
- Preceded_by -> Change data into IDs
Action items
.ace file to modify located here: /home/acedb/draciti/Life_stage and is named Life_stageWS230.ace
Modify the .ace dump as follows
1) Object should be replaced with ID instead of name using the mapping table
2) A new tag called Public_name should be added and should contain the name that was previously the object name
3) the following tags should be replaced with ID instead of name
- Contained_in
- Followed_by
- Preceded_by
- Sub_stage
4) The lines containing Remark "goid: WBls:#######" should be deleted
Mapping table is here
http://tazendra.caltech.edu/~azurebrd/var/work/lifestage
script at : /home/acedb/draciti/Life_stage/parse.pl output goes to script. Errors from lack of mappings are in the file. current output in file "out"
the script that updates the OA runs at 8pm every day and is at
/home/postgres/work/pgpopulation/obo_oa_ontologies/update_obo_oa_ontologies.pl
gets the data from here https://raw.github.com/draciti/Life-stage-obo/master/worm_development.obo
The script runs when the date in the obo file changes.
Deleting terms
In WS230 the following terms were used in Citace Minus but were not present in the obo file:
L1 Larva Male L2 Larva male L3 Larva male
the last term generated was L4 Larva male:
[Term] id: WBls:0000073 name: L4 larva male def: "The fourth stage larva male. At 25 Centigrade\, it ranges 40-49.5 hours after fertilization\, 26-35.5 hours after hatch." [wb:wjc] is_a: WBls:0000038 ! L4 larva
Raymond suggested to leave the sex distinction out of the ontology. We are therefore NOT adding these new terms. Checked in the Life stage WS230 dump -> no data associated with these terms, we could delete them from the obo file and modify citace minus accordingly. Waiting for Wen's feedback -> OK. Deleted the 3 L1 Larva Male, L2 Larva male, L3 Larva male, L4 larva hermaphrodite, mixed, mixed stage terms from Citace Minus
Deposited on spica (Data for CitaceMinus) the file Life_stageD with the corrections and the Name_to_ID change. (05152012DR) Create terms for one cell embryo and oocyte?
Adding terms
Added 11-15 days post-L4 adult hermaphrodite using obo edit as per Wen Chen request (WBPaper38462).
id: WBls:0000074 name: 11-15 days post-L4 adult hermaphrodite def: "At 20 Centigrade\\: 11-15 days after L4-adult molt. 14-18 days after first cleavage." [wb:dr] is_a: WBls:0000057 ! adult hermaphrodite created_by: danielaraciti creation_date: 2012-05-22T14:35:04Z
Aligning worm_development_obo with acedb
Daniela will hand over the Life_stage ontology to Raymond (February 2019) As per their discussion there will be no model changes to align the worm_development_obo file with acedb. The only change was to add A UNIQUE ans #Evidence hash in the Definition, in order to capture the paper evidence.
1) Is_a and part_of relationships Is_a and Part_of relationships will both convert into Contained_in 2) Preceded_by and Immediately_preceded_by relationships Preceded_by and Immediately_preceded_by relationships will both convert into Preceded_by 3) Synonym will convert into Other_name See the section: 'Added synonyms in the obo file' in this wiki (March 21st 2017) 4) Comment will convert into Remark
These lines we will –D on CitaceMinus, as they will come from the obo file converted into .ace: Public_name Sub_stage ?Life_stage XREF Contained_in Contained_in ?Life_stage XREF Sub_stage Preceded_by ?Life_stage XREF Followed_by Followed_by ?Life_stage XREF Preceded_by Remark ?Text #Evidence Definition ?Text Other_name ?Text // added for Wen [020607 krb]
conversion from obo to .ace
Added preceded_by relationships in the obo file using the dump "LS_Preceded_by.ace" provided by Wen -DR. Added additional preceded_by relationships that were not in the file-DR. The file is located on Lario here /Users/danielaraciti/Desktop/Life_stage/Relationships.
The conversion from obo to .ace should be as follows
OBO id: WBls:0000007 name: 2-cell embryo def: "0-20min after first cleavage at 20 Centigrade. Contains 2 cells." [wb:wjc] is_a: WBls:0000005 ! blastula embryo relationship: preceded_by WBls:0000006 ! 1-cell embryo .ace Life_stage : "WBls:0000007" Public_name "2-cell embryo" Definition "0-20min after first cleavage at 20 Centigrade. Contains 2 cells." Contained_in "WBls:0000005" Preceded_by "WBls:0000006"
- script at :
/home/acedb/draciti/Life_stage/lifestageAceFromObo.pl
- generates :
/home/acedb/draciti/Life_stage/lifestage.ace
Here's the relevant matching :
if ($obj =~ m/id: (WBls:\d+)/) { $id = $1;
$ace .= qq(Life_stage : "$id"\n);
if ($obj =~ m/name: (.*)/) { $ace .= qq(Public_name\t"$1"\n); }
if ($obj =~ m/def: "(.*)"/) { $ace .= qq(Definition\t"$1"\n); }
if ($obj =~ m/is_a: (WBls:\d+)/) { $ace .= qq(Contained_in\t"$1"\n); }
if ($obj =~ m/relationship: preceded_by (WBls:\d+)/) { $ace .=
qq(Preceded_by\t"$1"\n); }
$ace .= qq(\n);
meaning :
- the id is the WBls
- the name is everything
- the def is what's in quotes
- the is_a is the WBls
- the relationship is the WBls and it must say "relationship: precededby WBls:\d+"
It always downloads the .obo file from the URL https://raw.github.com/draciti/Life-stage-obo/master/worm_development.obo, and always regenerates the .ace file.
Broad synonyms
Ref Issue #72 on Textpresso github Nematoda Life Stage Category - Term Names not Optimal for Searching #72
Juancarlos automatically scripted in a broad synonym for all the terms that had Ce as suffix, so to have the life stages more amenable for textpresso searches.
Script here:/home/acedb/draciti/Life_stage/Broad_synonym/parse_worm_development_obo.pl
output:worm_development.obo.out
Committed on Git on July 13th 2016
Added synonyms in the obo file
Since we are going to dump all the ontology data from the obo file and convert into .ace, we will -D the Other_name lines on Citace minus, need to transfer the missing synonyms into the obo:
Added synonyms in the obo file, 9 instances they were listed in the .ace as Other_name 1) Life_stage : "WBls:0000006" Public_name "1-cell embryo Ce" Other_name "zygote" 2) Life_stage : "WBls:0000011" Public_name "51-cell embryo Ce" Other_name "50-70 cell embryo Ce" 3) Life_stage : "WBls:0000012" Public_name "88-cell embryo Ce" Other_name "90 cell embryo Ce" Did not add it as synonym in the obo as they seem to be 2 different stages 4) Life_stage : "WBls:0000020” Public_name "3-fold embryo Ce" Other_name "pretzel embryo" 5) Life_stage : "WBls:0000021” Public_name "fully-elongated embryo Ce" Other_name "pre-hatched embryo Ce" other synonyms were already accounted for in the .ace
-D Citace Minus
Citace minus dumps back ups here: Users/danielaraciti/Desktop/Life-stage-obo/Citace minus back ups February 2019
Discuss with Wen what is best to do for the -D file to submit to CitaceMinus. Daniela dumped the Life stage class from citace (file life_stage111213.ace) and -D all the lines that were present in the lifestage.ace generated by Juancarlos' script. All files are on lario here: /Users/danielaraciti/Desktop/Life_stage/DashD_november_2013 (life_stage111213.ace, lifestage.ace, and DashD_life_stage111213.ace)
If you decide to move away from citace minus completely, remark in Citace Minus could be stored in Comment in OBO edit and the comment in OBO edit should then be dumped as remark from Juancarlos' script. Same thing for othername, could go into synonyms and then dumped as other name.
One option would be to remove the whole class from citace minus. In this way we can rely just on the obo file and run the script that converts into .ace, then upload the lifestage.ace on citace at each upload.
In order to do that we should check that all tags are being converted in the .ace file
- Definition -> in OBO file
- Public_name -> in OBO file
- Contained_in -> in OBO file
- Preceded_by -> in OBO file
- Followed_by -> populated via acedb
- Sub_stage -> populated via acedb
- Anatomy_term -> populated through the Anatomy_term class?
- Reference -> populated how?
- Expr_pattern -> populated through the Expression pattern class?
- Cell_group is this coming from the cell group class?
- Cell is this coming from the cell class?
There are few tags not accounted in the obo file (Anatomy_term, Reference, Cell_group, Cell). So we have decided to keep the class in citace minus and add-remove as needed.
Synonym -> Other_name synonym: "" should convert in Other_name ""
Comment -> Remark comment: should convert in Remark ""
part_of should convert into contained_in starts_at_the_end_of should convert into preceded_by
some of the changes to apply to the script if we decide to delete the whole class are:
I need to modify an existing script
the script is here:
/home/acedb/draciti/Life_stage/lifestageAceFromObo.pl*
and generates /home/acedb/draciti/Life_stage/lifestage.ace
it takes data from this obo file:
https://raw.github.com/draciti/Life-stage-obo/master/worm_development.obo
and creates an .ace.
similarly as
if ($obj =~ m/is_a: (WBls:\d+)/) { $ace .= qq(Contained_in\t"$1"\n); }
and starts_at_the_end_of should convert into Immediately_preceded_by
similarly as:
if ($obj =~ m/relationship: starts_at_the_end_of (WBls:\d+)/) { $ace .=
qq(Immediately_receded_by\t"$1"\n); }
The comment field should be dumped as remark
Comment -> Remark
comment:
should convert in
Remark ""
and synonym into Other_name
Synonym -> Other_name
synonym: ""
should convert in
Other_name ""
Done-> See the section: Added synonyms in the obo file in this wiki
alt_IDs
There are only 2 entry that have alt_IDs in the obo file:
[Term] id: WBls:0000077 name: Brugia sheathed microfilaria alt_id: WBls:0000662 and [Term] id: WBls:0000663 name: Brugia unsheathed microfilaria alt_id: WBls:0000078
Only Expr11949 was using the term WBls:0000078: changed in WBls:0000663 in expression_OA
Gene association file for Life_stage
Specifications for a GAF for gene-life_stage data
other species
generic terms for other species -i.e. non C.elegans, non B. Malayi
Life_stage : "WBls:0000101" Public_name "all stages" Definition "All developmental stages, including embryo, larva and adult stage." Contained_in "WBls:0000075" Life_stage : "WBls:0000102" Public_name "embryo" Definition "The whole period of embryogenesis, from the formation of an egg till its hatch." Contained_in "WBls:0000101" Life_stage : "WBls:0000103" Public_name "postembryonic" Definition "The stage after hatch till death." Contained_in "WBls:0000101" Preceded_by "WBls:0000102" Life_stage : "WBls:0000104" Public_name "adult" Definition "The stage after an animal is fully-developed and reaches maturity." Contained_in "WBls:0000103" Preceded_by "WBls:0000105" Life_stage : "WBls:0000105" Public_name "larva" Definition "From the time after hatch till becomes adult." Contained_in "WBls:0000103" Life_stage : "WBls:0000106" Public_name "L1 larva" Definition "The first stage larva." Contained_in "WBls:0000105" Preceded_by "WBls:0000110" Life_stage : "WBls:0000107" Public_name "L2 larva" Definition "The second stage larva." Contained_in "WBls:0000105" Preceded_by "WBls:0000106" Life_stage : "WBls:0000108" Public_name "L3 larva" Definition "The third stage larva." Contained_in "WBls:0000105" Preceded_by "WBls:0000107" Life_stage : "WBls:0000109" Public_name "L4 larva" Definition "The fourth stage larva." Contained_in "WBls:0000105" Preceded_by "WBls:0000108" Life_stage : "WBls:0000110" Public_name "sheathed microfilaria" Definition "Microfilaria is the stage which develops from the egg and precedes the L1 stage. The larvae are surrounded by a membrane thought to be derived from the vitelline membrane." Contained_in "WBls:0000105"
Further additions
please list below the life stages that have been described or used in the literature for other species. This list will be periodically revised in order to expand the life stage ontology. If there is a formal description of the life stage i.e. definition of "sheathed microfilaria", "enclosing embryo" etc.. please include it with a reference (WBPaperID).
- C. briggsae
- ...
- C. japonica
- ...
- C. remanei
- ...
- C. sp11
- ...
- P. pacificus
- ...
- A. suum
- ...