Gene Interaction
'links to relevant pages
Caltech documentation
Contents
- 1 Interaction Curation
- 2 Pipeline
- 3 Interaction Models
- 4 Gene_gene Interaction OA
- 5 Some Notes for gene_gene_interaction
- 6 The new Interaction OA, March 2012
- 7 To go live on tazendra
- 8 Instructions on How to Use the New OA
- 9 Nightly Cron Job to Assign New Interaction IDs
- 10 Interaction OA .ACE Dumper
Interaction Curation
Pipeline
semi-automatic curation with textpresso extracted sentences
- There are 2138 sentences (actually 2133 sentences) in the sourcefile /home/postgres/work/pgpopulation/genegeneinteraction/20091002-xiaodong/ggi_20091002
- paper starts at WBPaper00028425, ends at WBPaper00035225
- .ace dumper at /home/acedb/xiaodong/gene_gene_interaction/dump_ggi_ace.pl
- go to the directory and do: ./dump_ggi_ace.pl > some_file.ace
- Populate textpresso data in tazendra OA: done on 20110110 -X**
- cd to directory on tazendra: /home/acedb/xiaodong/textpresso_ggi
- mkdir directionay_name (eg 20110106)
- cd directory_name (eg 20110106)
- get Arun's result file (35225-35725.txt in the directory)
- run script: ./populate_textpresso_ggi_to_OA.pl 20110106/35225-35725.txt WBPerson4793 > 20110106/35225-35725.pg (with first argument file_name as input file, and second argument WBPersonID, then output file)
- after running, '20110106/35225-35725.pg' should be in '20111106' directory.
upload Gary and Chris RNAi-based interaction objects into OA
- Reading file created by Igor's script into aceDB
- you can use the empty database by ssh -X citpub@spica.caltech.edu
- then cd CitaceMirror
- then type 'ts' to launch an empty acedb
- Dumping no-worry .ace file
- Then parse into OA
- scp ace file to same directory as below
- ssh acedb@tazendra.caltech.edu (directory xiaodong/interaction_ace_parsing)
- To run : ./parse_ace_interaction_oa.pl gary_RNAi.ace WBPerson1823
- where gary_RNAi.ace is the <inputfile>,the first argument, and WBPerson# is the second argument.
dump .ace file from OA for upload
- on tazendra: /home/acedb/xiaodong/oa_interactions_dumper
- run script by calling: ./use_package.pl
- error file will be spitted out in the same directory after every run. inform curators to check the errors.
- Old dumper outputs (err* and interaction* files) will now be archived in the following directory:
- /home/acedb/xiaodong/oa_interactions_dumper/Interaction_OA_Dumper_Output_Archive -- CG 10-31-2012
Interaction Models
Current Models
The current ?Interaction model now consolidates ?Gene_regulation objects, ?YH objects, and ?Interaction objects into a single class, ?Interaction. The proposed ?Physical_interaction model has also been consolidated into this larger model. Note that #Interaction_info and #Interaction_type have been deprecated.
?Interaction Interaction_type Physical
Predicted
Regulatory Change_of_localization // Indicates regulation of localization
Change_of_expression_level // Indicates regulation of expression level (RNA, protein)
Genetic Genetic_interaction // Indicates a generic genetic interaction that may not be accurately captured by any other term
Negative_genetic // General case in which one genetic perturbation exacerbates the effects of second perturbation
Synthetic // Two genetic perturbations are individually wild type but produce a phenotype when combined
Enhancement // One genetic perturbation exacerbates the effects of second perturbation
Unilateral_enhancement // One genetic perturbation exacerbates the effects of a second perturbation, which is otherwise wild type
Mutual_enhancement // Two genetic perturbations individually result in a phenotype and combine to result in a more severe phenotype than either individual perturbation
Suppression // One genetic perturbation suppresses the effects of second perturbation
Unilateral_suppression // One genetic perturbation suppresses the effects of second perturbation, which is otherwise wild type
Mutual_suppression // Two genetic perturbations individually result in a phenotype and combine to result in a less severe phenotype than either individual perturbation
Asynthetic // Two genetic perturbations individually result in an identical phenotype which is also identical to the phenotype of their combination
Suppression_enhancement // A double genetic perturbation yields a phenotype intermediate to that of either individual perturbation
Epistasis // The phenotype of one genetic perturbation masks the phenotype of a second perturbation
Maximal_epistasis // The more severe phenotype exhibited by two genetic perturbations is observed when both perturbations are combined
Minimal_epistasis // The less severe phenotype exhibited by two genetic perturbations is observed when both perturbations are combined
Suppression_epistasis // One genetic perturbation results in a phenotype which is suppressed back to wild type when combined with a second (wild type) perturbation
Agonistic_epistasis // Combined phenotype is identical to the the single perturbation which is closer to the expected phenotype as determined by the neutrality function
Antagonistic_epistasis // Two genetic perturbations each result in opposite phenotypes and the combined phenotype is identical to the the single perturbation which is furthest from the expected phenotype as determined by the neutrality function
Oversuppression // One genetic perturbation suppresses the phenotype of a second perturbation beyond wild type (producing an opposite phenotype)
Unilateral_oversuppression // One genetic perturbation suppresses the phenotype of a second (wild type) perturbation beyond wild type (producing an opposite phenotype)
Mutual_oversuppression // Two genetic perturbations individually result in a similar phenotype but result in an opposite phenotype when combined
Complex_oversuppression // Two genetic perturbations each result in opposite phenotypes and the combined phenotype is (relative to expectation) suppressed beyond wild type, resulting in a phenotype opposite to that expected
Oversuppression_enhancement // Two genetic perturbations each result in opposite phenotypes and the combined phenotype is oversuppressed relative to one perturbation and enhanced relative to the other perturbation
Phenotype_bias // " is less severe than either original phenotype, but deviates from expectation
Biased_suppression // Two genetic perturbations each result in opposite phenotypes and the combined phenotype is less severe than either original phenotype, and less severe than expected
Biased_enhancement // Two genetic perturbations each result in opposite phenotypes and the combined phenotype is less severe than either original phenotype, but more severe than expected
Complex_phenotype_bias // Two genetic perturbations each result in opposite phenotypes, and although the combined phenotype is expected to be wild type, the actual combined perturbations result in a phenotype less severe than either original phenotype
No_interaction // Negative data; no interaction was observed after testing
Interactor PCR_interactor ?PCR_product #Interactor_info // PCR_product of the interacting gene or protein, e.g. Yeast Two Hybrid experiments
Sequence_interactor ?Sequence #Interactor_info // Sequence of the interacting gene or protein
Interactor_overlapping_CDS ?CDS #Interactor_info // CDS of the interacting gene or protein (or related sequence)
Interactor_overlapping_gene ?Gene XREF Interaction #Interactor_info // Gene (or portion of gene) involved in the interaction
Interactor_overlapping_protein ?Protein XREF Interaction #Interactor_info // Protein (or portion of protein) involved in the interaction
Rearrangement ?Rearrangement XREF Interactor #Interactor_info // Rearrangement involved in the interaction
Molecule_regulator ?Molecule XREF Interaction #Interactor_info // Molecule that regulates a gene or protein (ported from Gene_regulation class)
Other_regulator ?Text #Interactor_info // Free text describing a regulator entity or condition that does not fall into a standard WormBase category
Other_regulated ?Text #Interactor_info // Free text describing a regulated entity or condition that does not fall into a standard WormBase category
Interaction_summary ?Text #Evidence
Detection_method Affinity_capture_luminescence // A physical interaction detection technique
Affinity_capture_MS // A physical interaction detection technique
Affinity_capture_RNA // A physical interaction detection technique
Affinity_capture_Western // A physical interaction detection technique
Cofractionation // A physical interaction detection technique
Colocalization // A physical interaction detection technique
Copurification // A physical interaction detection technique
Fluorescence_resonance_energy_transfer // A physical interaction detection technique
Protein_fragment_complementation_assay // A physical interaction detection technique
Yeast_two_hybrid // A physical interaction detection technique (Protein-protein)
Biochemical_activity // A physical interaction detection technique
Cocrystal_structure // A physical interaction detection technique
Far_western // A physical interaction detection technique
Protein_peptide // A physical interaction detection technique
Protein_RNA // A physical interaction detection technique
Reconstituted_complex // A physical interaction detection technique
Yeast_one_hybrid // A physical interaction detection technique (Protein-DNA)
Directed_yeast_one_hybrid // A physical interaction detection technique (Protein-DNA)
Antibody // A regulatory interaction detection technique; Antibody name and details captured in Interactor_info hash
Reporter_gene ?Text // A regulatory interaction detection technique
Transgene // A regulatory interaction detection technique; Trasnsgene name and details captured in Interactor_info hash
In_situ Text // A regulatory interaction detection technique
Northern Text // A regulatory interaction detection technique
Western Text // A regulatory interaction detection technique
RT_PCR Text // A regulatory interaction detection technique
Other_method ?Text // A regulatory interaction detection technique
//Physical interaction-specific tag
Library_info Library_screened Text INT // In the context of Yeast Two Hybrid or Yeast One Hybrid screens, for example, the library may have been cDNA library or some other pool of clones
Origin From_laboratory UNIQUE ?Laboratory // A library generated at an academic laboratory
From_company UNIQUE ?Text // A library generated at a company
//Genetic interaction-specific tags
Deviation_from_expectation Text // A text description of the way in which the phenotype deviated from expectation in genetic interactions
Neutrality_function UNIQUE Multiplicative // The multiplicative neutrality function defines expectation as the product of two quantified phenotypes (relative to wild type)
Additive // The additive neutrality function defines expectation as the sum of two quantified phenotypes (relative to wild type)
Minimal // The minimal neutrality function defines expectation as the most severe of two quantified phenotypes (relative to wild type)
//Gene regulation-specific tags
Regulation_level Transcriptional // Regulation occurs at the transcriptional level
Post_transcriptional // Regulation occurs at the post-transcriptional level
Post_translational // Regulation occurs at the post-translational level
Interaction_associated_feature ?Feature XREF Associated_with_gene_regulation #Evidence //to curate sequence feature connection [ar2]
Regulation_result Positive_regulate #GR_condition
Negative_regulate #GR_condition
Does_not_regulate #GR_condition // added to capture negative data [040220 krb]
//General tags
Confidence Description Text // Free text description of the confidence, e.g. "Core" vs "Noncore" (Vidal Interactome terms)
P_value UNIQUE Float // P-value confidence of interaction, if given
Log_likelihood_score UNIQUE Float // Only used for Predicted interactions
Throughput UNIQUE High_throughput //See BioGRID curation criteria for discussion: http://www.yeastgenome.org/help/BiogridCuration.html
Low_throughput
Interaction_RNAi ?RNAi XREF Interaction // RNAi experiment associated with the interaction
Interaction_phenotype ?Phenotype XREF Interaction // Phenotype associated with a genetic interaction
Unaffiliated_variation ?Variation // As opposed to Variations affiliated directly with an interacting gene, these are unaffiliated variations that somehow play a role in the interaction
Unaffiliated_transgene ?Transgene // As opposed to Transgenes affiliated directly with an interacting gene, these are unaffiliated Transgenes that somehow play a role in the interaction
Unaffiliated_antibody ?Antibody // As opposed to Antibodies affiliated directly with an interacting gene, these are unaffiliated Antibodies that somehow play a role in the interaction
Unaffiliated_expr_pattern ?Expr_pattern // As opposed to Expression Patterns affiliated directly with an interacting gene, these are unaffiliated Expression Patterns that somehow play a role in the interaction
Antibody_remark ?Text // Free text description of the antibody used to detect a regulation event //removed XREF from proposal
WBProcess ?WBProcess XREF Interaction // WormBase biological process associated with the interaction
DB_info Database ?Database ?Database_field ?Accession_number // Any database reference to the interaction outside of WormBase, e.g. BioGRID, Interactome
Paper ?Paper XREF Interaction
Remark ?Text #Evidence
#Interactor_info Interactor_type Non_directional // An interactor that has no inherent directionality Bait // The interactor of interest or focus; the focus/starting point of an interaction screen Target // The discovered interactor; interactors found as a result of an interaction screen Effector // In a genetic interaction, the perturbation that affects the phenotype of the other perturbation Affected // In a genetic interaction, the perturbation whose phenotype is affected by the other perturbation Trans_regulator // A trans-acting regulator, e.g. a transcription factor Cis_regulator // A cis-acting regulator, e.g. an enhancer element Trans_regulated // A gene regulated in trans, e.g. by a transcription factor Cis_regulated // A gene regulated in cis, e.g. by an enhancer element Expr_pattern ?Expr_pattern // An expression pattern altered to indicate Variation ?Variation XREF Interactor // (allele, polymorphism, etc.) involved in the interaction //removed XREF from proposal Intragenic_effector_variation ?Variation XREF Interactor // An effector variation that interacts with another variation from the same gene Intragenic_affected_variation ?Variation XREF Interactor // An affected variation that interacts with another variation from the same gene Transgene ?Transgene XREF Interactor // Transgene XREF Interactor that carries an interacting gene //removed XREF from proposal Antibody ?Antibody XREF Interactor // Antibody used to detect a regulation event Remark ?Text #Evidence // Info about reagents that the model can't capture (e.g. co_suppression, RNA_reagent, etc.)
In the adoption of this new ?Interaction model in WS231, we have consolidated the ?YH and ?Gene_regulation class into the ?Interaction class. As of WS231, there were 3831 ?Gene_regulation objects, which were then converted to ?Interaction objects with IDs WBInteraction000501384 through WBInteraction000505214. As of WS231, there were 11,993 ?YH objects, which were then converted to ?Interaction objects with IDs WBInteraction000505215 through WBInteraction000517207.
Old Models
?Interaction Evidence #Evidence
Interactor ?Gene XREF Interaction #Interactor_info
Interaction_type Genetic #Interaction_info
Regulatory #Interaction_info
No_interaction #Interaction_info
Predicted_interaction #Interaction_info
Physical_interaction #Interaction_info
Suppression #Interaction_info
Enhancement #Interaction_info
Synthetic #Interaction_info
Epistasis #Interaction_info
Mutual_enhancement #Interaction_info
Mutual_suppression #Interaction_info
Confidence Confidence_level UNIQUE Float
P_value UNIQUE Float
Log_likelihood_score UNIQUE Float
Paper ?Paper XREF Interaction
DB_info Database ?Database ?Database_field ?Accession_number
Remark ?Text #Evidence
#Interactor_info
Variation ?Variation XREF Interactor
Transgene ?Transgene XREF Interactor
Remark ?Text #Evidence //info about the reagents that the model can't capture goes here (e.g. co_suppression, RNA_reagent, etc.)
#Interaction_info
Interaction_RNAi ?RNAi XREF Interaction
Effector ?Gene //master, upstream
Effected ?Gene //subject, downstream
Non_directional ?Gene //e.g. synthetic interactions - Igor
Interaction_phenotype ?Phenotype XREF Interaction
Confidence Confidence_level UNIQUE Float
P_value UNIQUE Float
#Interaction_type
Genetic //directional and non_directional
Physical_interaction
Regulation
No_interaction
Synthetic//non_directional
Epistasis
Enhancement
Suppression
Predicted //addition for WeiWei, non_direactional
Mutual_enhancement//non_directional
Mutual_suprression//non_directional
///////////////////////////////////////////////////////////////////////////////////
New Model Proposals
Physical Interaction Model v1.3
?Physical_interaction Evidence #Evidence
Interactor Non_directional_interactor PCR_non_directional_interactor UNIQUE ?PCR_product XREF to? ?Species
Sequence_non_directional_interactor UNIQUE ?Sequence XREF to? ?Species
Non_directional_interactor_overlapping_CDS ?CDS XREF to? ?Species #Evidence
Non_directional_interactor_overlapping_gene ?Gene XREF to ? ?Species #Evidence
Non_directional_interactor_DB_info ?Database ?Database_field UNIQUE ?Accession_number //BioGRID, BioGRIDID, Numerical Value
Bait PCR_bait UNIQUE ?PCR_product XREF to? ?Species
Sequence_bait UNIQUE ?Sequence XREF to? ?Species
Bait_overlapping_CDS ?CDS XREF to? ?Species #Evidence
Bait_overlapping_gene ?Gene XREF to? ?Species #Evidence
Bait_DB_info ?Database ?Database_field UNIQUE ?Accession_number //BioGRID, BioGRIDID, Numerical Value
Target PCR_target UNIQUE ?PCR_product XREF to? ?Species
Sequence_target UNIQUE ?Sequence XREF to? ?Species
Target_overlapping_CDS ?CDS XREF to? ?Species #Evidence
Target_overlapping_gene ?Gene XREF to? ?Species #Evidence
Target_DB_info ?Database ?Database_field UNIQUE ?Accession_number //BioGRID, BioGRIDID, Numerical Value
Experimental_System UNIQUE Affinity_capture-luminescence //Experimental_system includes WormBase tags values as well as BioGRID values
Affinity_capture-MS
Affinity_capture-RNA
Affinity_capture-Western
Co-fractionation
Co-localization
Co-purification
FRET
PCA
Two-hybrid
Biochemical_activity
Co-crystal_structure
Far_western
Protein_peptide
Protein_RNA
Reconstituted_complex
Y1H //BioGRID is not curating protein-DNA interactions. WB has both Y1H data and GO MF data.
Directed_Y1H Text
Protein_DNA
Throughput UNIQUE High_throughput //See BioGRID curation criteria for discussion: http://www.yeastgenome.org/help/BiogridCuration.html
Low_throughput
Library_info Library_screened UNIQUE ?Library //This could also just be ?Text. Doesn't look like ?Library class is used.
Origin From_laboratory UNIQUE ?Laboratory //XREF by making a Reagents tag in the ?Laboratory model?
From_company UNIQUE ?Text //We don't currently have a ?Company class. Should we?
Confidence ?Text //Not currently captured by BioGRID, but this tag can accommodate the legacy YH data.
Paper ?Paper XREF to ?
Remark ?Text #Evidence //How would remarks coming from BioGRID be attributed? Person_evidence? Curator_confirmed? Accession_evidence? Person or Curator would require a change to the dumping file from BioGRID.
Physical Interaction Model v1.2
The revised v1 includes: 1) a ?Species tag, 2) a slot to capture Non-directional interactors (for curating things like protein complexes purified over sedimentation gradients, i.e. where there is no clear Bait or Target directionality), and 3) change ?Confidence from a specific list of phrases or statistical methods to a ?Text tag since this information is expressed in many different ways in the literature so including specific text here doesn't seem practical. If we change this to ?Text, then I'd also remove the specific Interactome_core tag.
Also, current XREF tags in the ?YH model are YH_bait and YH_target. What would be a more appropriate name? Model below has Interaction_target, etc. but I think that's not clear enough. What about Physical_interaction_target?
Also, CDS and Gene, when overlapping, have #Evidence, but the PCR and Sequence do not. Why is this? Does it have to do with needing to indicate how a CDS or Gene was selected without a corresponding sequence?
?Physical_interaction Evidence #Evidence
Species UNIQUE ?Species
Interactor Non_directional_interactor PCR_non_directional_interactor UNIQUE ?PCR_product XREF ?
Sequence_non_directional_interactor UNIQUE ?Sequence XREF ?
Non_directional_interactor_overlapping_CDS ?CDS XREF ? #Evidence
Non_directional_interactor_overlapping_gene ?Gene XREF ? #Evidence
Bait PCR_bait UNIQUE ?PCR_product XREF ?
Sequence_bait UNIQUE ?Sequence XREF ?
Bait_overlapping_CDS ?CDS XREF ? #Evidence
Bait_overlapping_gene ?Gene XREF ? #Evidence
Target PCR_target UNIQUE ?PCR_product XREF ?
Sequence_target UNIQUE ?Sequence XREF ?
Target_overlapping_CDS ?CDS XREF ? #Evidence
Target_overlapping_gene ?Gene XREF ? #Evidence
Experiment_type Affinity_capture-luminescence
Affinity_capture-MS
Affinity_capture-RNA
Affinity_capture-Western
Co-fractionation
Co-localization
Co-purification
FRET
PCA
Two-hybrid
Biochemical_activity
Co-crystal_structure
Far_western
Protein_peptide
Protein_RNA
Reconstituted_complex
Y1H
Directed_Y1H Text
Protein_DNA
Throughput UNIQUE High_throughput //Need to define in context of physical interactions
Low_throughput //Same as above
Library_info Library UNIQUE ?Library //This could also just be ?Text. Doesn't look like ?Library class is used.
Origin From_laboratory UNIQUE ?Laboratory //XREF by making a Reagents tag in the ?Laboratory model?
From_company UNIQUE ?Text //We don't currently have a ?Company class. Should we?
Confidence ?Text //This can accommodate the great variety of language used to expressed this, if curated.
Paper ?Paper XREF Interaction //Should this XREF also be updated to Physical_interaction?
Remark ?Text #Evidence
Physical Interaction Model v1
This version of the model treats each instance of a physical interaction as a separate entity.
?Physical_interaction Evidence #Evidence
Interactor Bait PCR_bait UNIQUE ?PCR_product XREF Interaction_bait //Change XREF tag to Physical...
Sequence_bait UNIQUE ?Sequence XREF Interaction_bait
Bait_overlapping_CDS ?CDS XREF Interaction_bait #Evidence
Bait_overlapping_gene ?Gene XREF Interaction_bait #Evidence
Target PCR_target UNIQUE ?PCR_product XREF Interaction_target //Change Target to Hit? Also change XREF as above?
Sequence_target UNIQUE ?Sequence XREF Interaction_target
Target_overlapping_CDS ?CDS XREF Interaction_target #Evidence
Target_overlapping_gene ?Gene XREF Interaction_target #Evidence
Experiment_type Affinity_capture-luminescence
Affinity_capture-MS
Affinity_capture-RNA
Affinity_capture-Western
Co-fractionation
Co-localization
Co-purification
FRET
PCA
Two-hybrid
Biochemical_activity
Co-crystal_structure
Far_western
Protein_peptide
Protein_RNA
Reconstituted_complex
Y1H
Directed_Y1H Text
Protein_DNA
Throughput UNIQUE High_throughput //Need to define in context of physical interactions
Low_throughput //Same as above
Library_info Library UNIQUE ?Library //This could also just be ?Text. Doesn't look like ?Library class is used.
Origin Species UNIQUE ?Species
From_laboratory UNIQUE ?Laboratory //XREF by making a Reagents tag in the ?Laboratory model?
From_company UNIQUE ?Text //We don't currently have a ?Company class. Should we?
Confidence Confidence_level UNIQUE Float //Do we need this in the ?Physical_interaction model?
P_value UNIQUE Float //Same as above.
Log_likelihood_score UNIQUE Float //Same as above.
Interaction_frequency UNIQUE Int //This would hold the Int data in the existing Library_screened tag.
Interactome_type UNIQUE Interactome_core_1 //As defined in Li et al., 2004
Interactome_core_2
Interactome_core_3
Paper ?Paper XREF Interaction //Should this XREF also be updated to Physical_interaction?
Remark ?Text #Evidence
Physical Interaction Model v2
This version of the model gives a single interaction ID to two interacting entities, but each instance, or evidence for the interaction, is added in the #Interaction_info under the corresponding Experiment_type.
?Physical_interaction Evidence #Evidence
Interactor Bait PCR_bait UNIQUE ?PCR_product XREF Interaction_bait //Change XREF tag to Physical...
Sequence_bait UNIQUE ?Sequence XREF Interaction_bait
Bait_overlapping_CDS ?CDS XREF Interaction_bait #Evidence
Bait_overlapping_gene ?Gene XREF Interaction_bait #Evidence
Target PCR_target UNIQUE ?PCR_product XREF Interaction_target //Change Target to Hit? Also change XREF as above?
Sequence_target UNIQUE ?Sequence XREF Interaction_target
Target_overlapping_CDS ?CDS XREF Interaction_target #Evidence
Target_overlapping_gene ?Gene XREF Interaction_target #Evidence
Experiment_type Affinity_capture-luminescence #Interaction_info
Affinity_capture-MS #Interaction_info
Affinity_capture-RNA #Interaction_info
Affinity_capture-Western #Interaction_info
Co-fractionation #Interaction_info
Co-localization #Interaction_info
Co-purification #Interaction_info
FRET #Interaction_info
PCA #Interaction_info
Two-hybrid #Interaction_info
Biochemical_activity #Interaction_info
Co-crystal_structure #Interaction_info
Far_western #Interaction_info
Protein_peptide #Interaction_info
Protein_RNA #Interaction_info
Reconstituted_complex #Interaction_info
Y1H #Interaction_info
Directed_Y1H Text #Interaction_info
Protein_DNA #Interaction_info
Remark ?Text #Evidence
#Interaction_info Interaction_RNAi ?RNAi XREF Interaction
Effector ?Gene //master, upstream
Effected ?Gene //subject, downstream
Non_directional ?Gene //e.g. synthetic interactions - Igor
Interaction_phenotype ?Phenotype XREF Interaction
Throughput UNIQUE High_throughput
Low_throughput
Library_info Library UNIQUE ?Library
Origin Species UNIQUE ?Species
From_laboratory UNIQUE ?Laboratory
From_company ?Text
Confidence Confidence_level UNIQUE Float
P_value UNIQUE Float
Log_likelihood UNIQUE Float
Interaction_frequency UNIQUE Int
Interactome_type UNIQUE Interactome_core_1
Interactome_core_2
Interactome_noncore
Paper ?Paper XREF Interaction
Physical Interaction Model v3
This model keeps the physical interaction as part of the general ?Interaction model with the details again going into the #Interaction_info. The #Interaction_info would now contain the information about bait/hit directionality.
?Interaction Evidence #Evidence
Interactor ?Gene XREF Interaction #Interactor_info
Interaction_type Genetic #Interaction_info
Regulatory #Interaction_info
No_interaction #Interaction_info
Predicted_interaction #Interaction_info
Physical_interaction #Interaction_info
Suppression #Interaction_info
Enhancement #Interaction_info
Synthetic #Interaction_info
Epistasis #Interaction_info
Mutual_enhancement #Interaction_info
Mutual_suppression #Interaction_info
DB_info Database ?Database ?Database_field ?Accession_number
Remark ?Text #Evidence
#Interaction_info Interaction_RNAi ?RNAi XREF Interaction
Effector ?Gene //master, upstream
Effected ?Gene //subject, downstream
Bait PCR_bait UNIQUE ?PCR_product XREF Interaction_bait //Change XREF tag to Physical...
Sequence_bait UNIQUE ?Sequence XREF Interaction_bait
Bait_overlapping_CDS ?CDS XREF Interaction_bait #Evidence
Bait_overlapping_gene ?Gene XREF Interaction_bait #Evidence
Target PCR_target UNIQUE ?PCR_product XREF Interaction_target //Change Target to Hit? Also change XREF as above?
Sequence_target UNIQUE ?Sequence XREF Interaction_target
Target_overlapping_CDS ?CDS XREF Interaction_target #Evidence
Target_overlapping_gene ?Gene XREF Interaction_target #Evidence
Experiment_type Affinity_capture-luminescence
Affinity_capture-MS
Affinity_capture-RNA
Affinity_capture-Western
Co-fractionation
Co-localization
Co-purification
FRET
PCA
Two-hybrid
Biochemical_activity
Co-crystal_structure
Far_western
Protein_peptide
Protein_RNA
Reconstituted_complex
Y1H
Directed_Y1H Text
Protein_DNA
Throughput UNIQUE High_throughput
Low_throughput
Library_info Library UNIQUE ?Library //This could also just be ?Text. Doesn't look like ?Library class is used.
Origin Species UNIQUE ?Species
From_laboratory UNIQUE ?Laboratory //XREF by making a Reagents tag in the ?Laboratory model?
From_company UNIQUE ?Text //We don't currently have a ?Company class. Should we?
Confidence Confidence_level UNIQUE Float
P_value UNIQUE Float
Log_likelihood_score UNIQUE Float
Interaction_frequency UNIQUE Int //This would hold the Int data in the existing Library_screened tag.
Interactome_type UNIQUE Interactome_core_1 //As defined in Li et al., 2004
Interactome_core_2
Interactome_core_3
Paper ?Paper XREF Interaction //Should this XREF also be updated to Physical_interaction?
Remark ?Text #Evidence
Gene_gene Interaction OA
OA interface
- Tab 1
- PGID Same in new OA
- Interaction ID
- A new interaction ID is generated by clicking on 'new'. when 'duplicate', the ID from old entry will be in the field, but need to be deleted in order to get an new ID.
- Interaction ID will be assigned by cronjob daily at 4 am. This is for curators who would like to use 'Duplicate' to generate new objects with similar field entries and erase the Interaction IDs to let the cron job add new IDs overnight. The cronjob is at /home/acedb/xiaodong/assigning_interaction_ids/assign_interaction_ids.pl and we need to change it based on the new table structure -- J
- interaction field autocompletes now key off of the int_index table used by the interaction_ticket.cgi, it no longer keys off of the int_name table from this field in the interaction OA, which makes the rest of this line obsolete. (OBSOLETE If you mistakenly make a typo and assign a correct ID's value to some other ID, you will _not_ be able to bring it back (because it's an ontology) without going to postgres directly and editing the int_name and int_name_hst tables by pgid (in postgres called joinkey). You'll have to note the pgid and then manually change it in postgres. end obsolete)
- Non_directional In new OA, there will be a separate field for each interactor type: Gene, Sequence, CDS, PCR_Product, or Protein. Since "old" interaction objects only contain genes, any genes that are part of Interaction obejcts in which the Non_directional toggle was activated will need to move to the "Non-directional Gene(s)" field. if toggle is on, move all effector + effected genes to the new non-directional gene field. get rid of this field in new OA
- toggle OFF (default), means interaction is directional. there is effected/effector parties involve in the object.
- toggle ON (color change to red by click) means interaction is non_directional
- Interaction Type//dropdown list with 11 types showing in .ace template
- In the new OA, these 11 types will be mapped as follows:
- "Genetic" will become "Genetic - Genetic Interaction"
- "Regulatory" will remain "Regulatory"
- "No_interaction" will become "Genetic - No_interaction"
- "Predicted_interaction" will become "Predicted"
- "Physical_interaction" will become "Physical"
- "Suppression" will become "Genetic - Suppression"
- "Enhancement" will become "Genetic - Enhancement"
- "Synthetic" will become "Genetic - Synthetic"
- "Epistasis" will become "Genetic - Epistasis"
- "Mutual_enhancement" will become "Genetic - Mutual_enhancement"
- "Mutual_suppression" will become "Genetic - Mutual_suppression"
- Effected Gene //autocomplete WBGene, multiontology, corresponding to interactor in .ace file. order does not matter when dump to interactors
- In new OA, this field will be called "Affected Gene(s)" -- C If non-directional is on, move to non-directional gene, otherwise leave here
- Effected Variation //WBGene, WBVar, multiontology, autocomplete on Variation, store in separate lines ->.ace, Interactor "WBGene" Variation "WBVar"
- In new OA, this field will be called "Affected Variation(s)"; the dumping script will attempt to map these variations to their respetive genes and place the variation in that gene's interactor_info hash -- C
- In new OA, the gene(s) affiliated with this variation will become the "Affected Gene(s)" for this interaction.
- Effected Transgene_Name //ontology, autocomplete transgene object name, eg iaIs3.
- In new OA, all transgenes will go into the Transgene(s) field. merge both effector + effected.
- Effected Transgene_Gene // multi-ontology, autocomplete WBGene->.ace, Interactor "WBGene" Transgene "id". In case of multi genes, WBGene is followed by same transgene id. One wbgene for each .ace line ? Make sure you really want it this way, we can go with product/promoter if that's what you want, just make sure it's what you want. It matters having extra fields and scrolling and so forth. You'll see when the text fields become multi-ontology and ontology.
- In new OA, all transgenes will automatically be mapped to their associated genes based on all genes in the transgene OA's gene + driven_by_gene field. remove this field.
- In new OA, genes from the "Effected Transgene Gene" field will move to the "Affected Gene" field.
- Effected Other Type //dropdown list of 'Chemicals' and 'Transgene'
- In new OA, these will be named "Affected Other Type" and dump as remark.
- Effected Other //free text field now, however, when entering chemicals make sure to enter common names followed by mesh IDs in parenthesis for later ontologinization.
- In new OA, these will be named "Affected Other" and dump as remark.
- Effector Gene //autocomplete WBGene, multiontology. corresponding to interactor in .ace file. order does not matter when dump to interactors
- In new OA, these genes will all go in the "Effector Gene(s)" field. If non-directional is on, move to non-directional gene, otherwise leave here
- Effector Variation //WBGene, WBVar, autocomplete multiontology on variation, store in separate lines ->.ace, Interactor "WBGene" Variation "WBVar". use name server to map variation to gene, or the file Karen gave you to map variation to gene for variation OA.
- In new OA, this field will be the same; the dumping script will attempt to map these variations to their respetive genes and place the variation in that gene's interactor_info hash -- C
- In new OA, the gene(s) affiliated with this variation will become the "Effector Gene(s)" for this interaction.
- Effector Transgene_Name //autocomplete name, ontology
- In new OA, all transgenes will go into the Transgene(s) field. merge both effector + effected.
- Effector Transgene_Gene // autocomplete WBGene, multi-ontology, ->.ace, Interactor "WBGene" Transgene "id". In case of multi genes, WBGene is followed by same transgene id.
- In new OA, all transgenes will automatically be mapped to their associated genes based on all genes in the transgene OA's gene + driven_by_gene field. remove this field.
- In new OA, genes from the "Effector Transgene Gene" field will move to the "Effector Gene" field.
- Effector Other Type //dropdown list of 'Chemicals' and 'Transgene'
- In new OA, these will stay the same and dump as Remark
- Effector Other //free text field
- two fields above will be dumped in remark field.
- In new OA, these will stay the same and dump as Remark
- Effected Gene //autocomplete WBGene, multiontology, corresponding to interactor in .ace file. order does not matter when dump to interactors
Note: Gene, Variation, and Transgene_Gene all refer to different genes. There is no pairing problem.
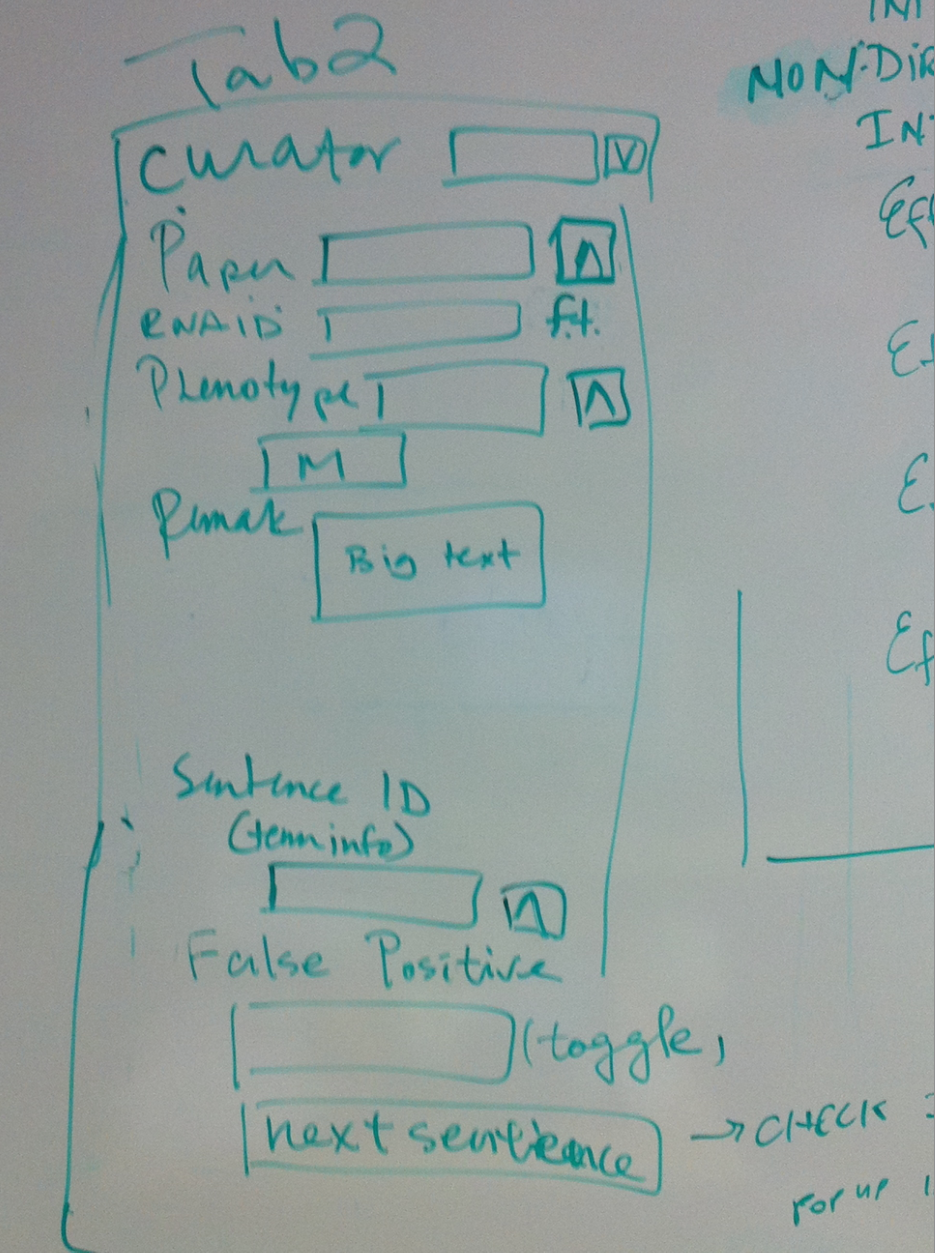
- Tab 2
- Curator//dropdown list Same in new OA
- Paper//ontology Same in new OA
- RNAi ID//free text field In new OA, this RNAi object will fill the "RNAi" field if it is not from WBPaper00029258 (hence large scale); if it is from WBPaper00029258, this will go in the "Large scale RNAi" field as free text
- Phenotype//multiontology In new OA, this Phenotype object will fill the "Interaction phenotype" field. keep OA field the same, change dumper.
- Remark//big text Same in new OA
- Sentence ID//sentence shows in term info Same in new OA
- False Positive//toggle, will not give an id or no dump if the sentence is false positive, containing no interaction info Same in new OA
Postgres Table Names
'Interaction ID' = int_name
'Curator' = int_curator
'Process' = int_process
'Database Field Accession Number' = int_database
'Paper' = int_paper
'Interaction Type' = int_type
'Interaction Summary' = int_summary
'Remark' = int_remark
'Physical interaction detection method' = int_detectionmethod
'Library screened and times found' = int_library
'From Laboratory' = int_laboratory
'From Company' = int_company
'PCR Bait' = int_pcrbait
'PCR Target(s)' = int_pcrtarget
'Non-directional PCR(s) = int_pcrnondir
'Sequence Bait' = int_sequencebait
'Sequence Target(s)' = int_sequencetarget
'Non-directional Sequence(s)' = int_sequencenondir
'Bait overlapping CDS' = int_cdsbait
'Target overlapping CDS(s)' = int_cdstarget
'Non-directional overlapping CDS(s)' = int_cdsnondir
'Bait overlapping protein' = int_proteinbait
'Target overlapping protein' = int_proteintarget
'Non-directional overlapping protein' = int_proteinnondir
'Bait overlapping gene' = int_genebait
'Target overlapping gene' = int_genetarget
'Antibody' = int_antibody
'Antibody remark' = int_antibodyremark
'Non-directional Gene(s)' = int_genenondir
'Effector Gene(s)' = int_geneone
'Affected Gene(s)' = int_genetwo
'Non-directional Rearrangement(s)' = int_rearrnondir
'Effector Rearrangement(s)' = int_rearrone
'Affected Rearrangement(s)' = int_rearrtwo
'Effector Other Type' = int_otheronetype
'Effector Other' = int_otherone
'Affected Other Type' = int_othertwotype
'Affected Other' = int_othertwo
'Deviation from expectation' = int_deviation
'Neutrality function' = int_neutralityfxn
'Large scale RNAi' = int_lsrnai
'RNAi' = int_rnai
'Interaction Phenotype(s)' = int_phenotype
'Expression pattern(s)' = int_exprpattern
'Non-directional Variation(s)' = int_variationnondir
'Effector Variation(s)' = int_variationone
'Affected Variation(s)' = int_variationtwo
'Intragenic Effector Variation(s)' = int_intravariationone
'Intragenic Affected Variation(s)' = int_intravariationtwo
'Transgene(s)' = int_transgene
'Person' = int_person
'Confidence description' = int_confidence
'P-value' = int_pvalue
'Log-likelihood score' = int_loglikelihood
'High_hroughput' = int_throughput
'Sentence ID' = int_sentid
'False Positive' = int_falsepositive
Old Table Names
'Non_directional' = int_nondirectional
'Effector Transgene Name' = int_transgeneone
'Effector Transgene Gene' = int_transgeneonegene
'Affected Transgene Name' = int_transgenetwo
'Affected Transgene Gene' = int_transgenetwogene
.ace template: (OUTDATED)
- Interaction : ""
- Interactor "WBGene" Variation ""
- Interactor "WBGene" Transgene ""
- Interactor "WBGene"
- Interaction_type Genetic Effector ""
- Interaction_type Genetic Effected ""
- Interaction_type Genetic Non_directional ""
- Interaction_type Genetic Interaction_RNAi ""
- Interaction_type Genetic Interaction_phenotype ""
- Interaction_type Regulatory Effector ""
- Interaction_type Regulatory Effected ""
- Interaction_type Regulatory Non_directional ""
- Interaction_type Regulatory Interaction_RNAi ""
- Interaction_type Regulatory Interaction_phenotype ""
- Interaction_type No_interaction Effector ""
- Interaction_type No_interaction Effected ""
- Interaction_type No_interaction Non_directional ""
- Interaction_type No_interaction Interaction_RNAi ""
- Interaction_type No_interaction Interaction_phenotype ""
- Interaction_type Predicted_interaction Non_directional ""
- Interaction_type Predicted_interaction Interaction_RNAi ""
- Interaction_type Predicted_interaction Interaction_phenotype ""
- Interaction_type Physical_interaction Effector ""
- Interaction_type Physical_interaction Effected ""
- Interaction_type Physical_interaction Interaction_RNAi ""
- Interaction_type Physical_interaction Interaction_phenotype ""
- Interaction_type Suppression Effector ""
- Interaction_type Suppression Effected ""
- Interaction_type Suppression Interaction_RNAi ""
- Interaction_type Suppression Interaction_phenotype ""
- Interaction_type Enhancement Effector ""
- Interaction_type Enhancement Effected ""
- Interaction_type Enhancement Interaction_RNAi ""
- Interaction_type Enhancement Interaction_phenotype ""
- Interaction_type Synthetic Non_directional ""
- Interaction_type Synthetic Interaction_RNAi ""
- Interaction_type Synthetic Interaction_phenotype ""
- Interaction_type Epistasis Effector ""
- Interaction_type Epistasis Effected ""
- Interaction_type Epistasis Interaction_RNAi ""
- Interaction_type Epistasis Interaction_phenotype ""
- Interaction_type Mutual_enhancement Non_directional ""
- Interaction_type Mutual_enhancement Interaction_RNAi ""
- Interaction_type Mutual_enhancement Interaction_phenotype ""
- Interaction_type Mutual_suppression Non_directional ""
- Interaction_type Mutual_suppression Interaction_RNAi ""
- Interaction_type Mutual_suppression Interaction_phenotype ""
- Paper ""
- Remark ""
interaction objects source file
- there are 9242 interaction objects dumped from WS220 on Monday, 10/01/2010
- Juancarlos's parse results from this file:
/home/postgres/work/pgpopulation/interaction/20101004_xiaodong_start/out
There are two interactions in postgres, but not the .ace file : In postgres, no ace WBInteraction0008637 In postgres, no ace WBInteraction0008638//will be OA
There are 1290 interactions in .ace file not in postgres (so I imagine these are what we should read in ?)//these are RNAi based interaction objects, we want to include them in OA
There are >40000 interactions that have a ticket and are in neither .ace nor postgres.//these 398,619 interactions are from two large scale papers
Also there are interaction data in postgres without an interaction ID//will be assigned id from WBInteraction0500001.
Some Notes for gene_gene_interaction
Two Large Scale Interaction Data Sets
- Files and scripts for these large scale datasets have been moved into a new directory:
/home/acedb/xiaodong/oa_interactions_dumper/Large_Scale_Interactions
-- CG 10-31-2012
- WBPaper00027155 (Weiwei's science paper) has 23,128 objects, starting from WBInteraction000008637 and ending at WBInteraction000050578 (blank ids from WBInteraction000050579 to 000100000)
- Original .ACE file (in old .ACE interaction format) on Tazendra here:
/home/acedb/xiaodong/oa_interactions_dumper/Large_Scale_Interactions/Original_Files/27155_interaction.ace
- WBPaper00031465 (Lee's Nature Genetics paper) has 375,491 objects, starting from WBInteraction000100001, ending at WBInteraction000475491
- Original .ACE file (in old .ACE interaction format) on Tazendra here:
/home/acedb/xiaodong/oa_interactions_dumper/Large_Scale_Interactions/Original_Files/31465_interaction.ace
- The two large scale data sets have now been updated to the new interaction format (as of May 2013) and consolidated into a single file on Tazendra here:
/home/acedb/xiaodong/oa_interactions_dumper/Large_Scale_Interactions/Original_Large_Scale_Interactions_new_format.ace
The above file needs to be checked for dead genes before every upload by running the "historicGeneReplacementLSInteraction.pl" script in the same directory. Before running the script, make sure that the output file is appropriately named. The script prints all dead genes to the screen, so when running the script you may want to redirect the script output into a file so you can read the results later.
Outdated notes on large scale interaction files
ready_to_upload large scale data file: Large_scale_interactions_WS231.ace is located on tazendra: /home/acedb/xiaodong/oa_interactions_dumper should be uploaded for every uploadlarge scale .ace file needs to be checkd again dead genes for each upload using scripts in the same directory: find_invalid_genes_largescale.pl* - 08/31/2012 for upload WS234- before run, change source file (in file) to latest large scale .ace version (in scripts)
- dead/merged genes will appear on screen. replace merged genes, leave dead/dead genes for now
- change large scale.ace file name to latest upload version
then upload
- assigning_interaction_ids
- textpresso_ggi
- interaction_ace_parsing
- oa_interactions_dumper
Citace upload notes
- 2011.1.27
- caught-up at acedb reading: WBInteraction0500069 (Karen), new line in remark field. fixed in .ace file.
- some confusion on ids. found out ticket issuer was using sandbox data. fixed.
- 2011.5.4
- Karen needed to update variation_wbgene file on tazendra: /home/acedb/jolene/WS_AQL_queries/Variation_gene.txt
- Juancarlos changed the dumper to ignore line breaks and double spaces in remark field for dumping ace file
The new Interaction OA, March 2012
TAB1
- PGID Dumps as: N/A
- Interaction ID (Ontology) int_name Dumps as: Interaction: <Interaction ID>
- Curator - (Dropdown) int_curator Dumps as: N/A
- Process - ?WBProcess (MultiOntology) int_process Dumps as: WBProcess <WBProcess>
- Database, Field, & Accession Number - ?Database, Field, Accession_number (Free Text) int_database Dumps as: Database <Database> <Database_field> <Accession_number>
- For single entries, surround the Database, Field, and Accession number entries with double quotes and separate them with spaces like so: "Database" "Database Field" "Accession Number"
- If there are multiple entries, data to be entered like this: "Database 1" "Field 1" "Accession number 1" | "Database 2" "Field 2" "Accession number 2"
- Paper - ?Paper (Ontology) int_paper Dumps as: Paper <Paper>
- Interaction Type - Text (Multiple-Dropdown) int_type
- The options for Interaction Type will include:
- Physical Dumps as: Physical
- Predicted Dumps as: Predicted
- Genetic - Genetic interaction Dumps as: Genetic_interaction
- Genetic - Negative genetic Dumps as: Negative_genetic
- Genetic - Synthetic Dumps as: Synthetic
- Genetic - Enhancement Dumps as: Enhancement
- Genetic - Unilateral enhancement Dumps as: Unilateral_enhancement
- Genetic - Mutual enhancement Dumps as: Mutual_enhancement
- Genetic - Suppression Dumps as: Suppression
- Genetic - Complete suppression Dumps as: Complete_suppression NEW CG 9-3-2013
- Genetic - Partial suppression Dumps as: Partial_suppression NEW CG 9-3-2013
- Genetic - Unilateral suppression Dumps as: Unilateral_suppression
- Genetic - Complete unilateral suppression Dumps as: Complete_unilateral_suppression NEW CG 9-3-2013
- Genetic - Partial unilateral suppression Dumps as: Partial_unilateral_suppression NEW CG 9-3-2013
- Genetic - Mutual suppression Dumps as: Mutual_suppression
- Genetic - Complete mutual suppression Dumps as: Complete_mutual_suppression NEW CG 9-3-2013
- Genetic - Partial mutual suppression Dumps as: Partial_mutual_suppression NEW CG 9-3-2013
- Genetic - Asynthetic Dumps as: Asynthetic
- Genetic - Suppression/Enhancement Dumps as: Suppression_enhancement
- Genetic - Epistasis Dumps as: Epistasis
- Genetic - Positive epistasis Dumps as: Positive_epistasis NEW CG 9-3-2013
- Genetic - Maximal epistasis Dumps as: Maximal_epistasis
- Genetic - Minimal epistasis Dumps as: Minimal_epistasis
- Genetic - Neutral epistasis Dumps as: Neutral_epistasis NEW CG 9-3-2013
- Genetic - Qualitative epistasis Dumps as: Qualitative_epistasis NEW CG 9-3-2013
- Genetic - Opposing epistasis Dumps as: Opposing_epistasis NEW CG 9-3-2013
- Genetic - Quantitative epistasis Dumps as: Quantitative_epistasis NEW CG 9-3-2013
- The options for Interaction Type will include:
Genetic - Suppression/Epistasis Dumps as: Suppression_epistasisCG 9-3-2013Genetic - Agonistic epistasis Dumps as: Agonistic_epistasisCG 9-3-2013Genetic - Antagonistic epistasis Dumps as: Antagonistic_epistasisCG 9-3-2013
- Genetic - Neutral genetic Dumps as: Neutral_genetic NEW CG 9-3-2013
- Genetic - Oversuppression Dumps as: Oversuppression
- Genetic - Unilateral oversuppression Dumps as: Unilateral_oversuppression
- Genetic - Mutual oversuppression Dumps as: Mutual_oversuppression
Genetic - Complex oversuppression Dumps as: Complex_oversuppressionCG 9-3-2013- Genetic - Oversuppression/Enhancement Dumps as: Oversuppression_enhancement
- Genetic - Phenotype bias Dumps as: Phenotype_bias
Genetic - Biased suppression Dumps as: Biased_suppressionCG 9-3-2013Genetic - Biased enhancement Dumps as: Biased_enhancementCG 9-3-2013Genetic - Complex phenotype bias Dumps as: Complex_phenotype_biasCG 9-3-2013- Genetic - No interaction Dumps as: No_interaction
- Interaction Summary - bigtext int_summary Dumps as: Interaction_summary <Big_Text>
- Remark - bigtext int_remark Dumps as: Remark <Big_Text>
TAB2
- Physical interaction detection method (Multi-dropdown) int_detectionmethod
- The detection method options are:
- Affinity_capture_luminescence Dumps as: Affinity_capture_luminescence
- Affinity_capture_MS Dumps as: Affinity_capture_MS
- Affinity_capture_RNA Dumps as: Affinity_capture_RNA
- Affinity_capture_Western Dumps as: Affinity_capture_Western
- Cofractionation Dumps as: Cofractionation
- Colocalization Dumps as: Colocalization
- Copurification Dumps as: Copurification
- Fluorescence_resonance_energy_transfer Dumps as: Fluorescence_resonance_energy_transfer
- Protein_fragment_complementation_assay Dumps as: Protein_fragment_complementation_assay
- Yeast_two_hybrid Dumps as: Yeast_two_hybrid
- Biochemical_activity Dumps as: Biochemical_activity
- Cocrystal_structure Dumps as: Cocrystal_structure
- Far_western Dumps as: Far_western
- Protein_peptide Dumps as: Protein_peptide
- Protein_RNA Dumps as: Protein_RNA
- Reconstituted_complex Dumps as: Reconstituted_complex
- Yeast_one_hybrid Dumps as: Yeast_one_hybrid
- Directed_yeast_one_hybrid Dumps as: Directed_yeast_one_hybrid
- Electrophoretic_mobility_shift_assay_(EMSA) Dumps as: Electrophoretic_mobility_shift_assay_(EMSA) NEW CG 9-3-2013
- The detection method options are:
- Library screened/Times found - Text Text(Integer) int_library; separate multiple entries with pipes ('|') int_library Dumps as: Library_screened <Text> INT
- For single entries, surround the 'Library screened' entry with double quotes and separate the number with a space like so: "Library screened" 3
- For multiple entries, data should be entered as such: "Library screened 1" INT | "Library screened 2" INT
- From Laboratory - ?Laboratory (ontology) int_laboratory Dumps as: From_laboratory <Laboratory>
- From Company - Text; separate multiple entries with pipes ('|') int_company Dumps as: From_company <Text>
- PCR Bait - ?PCR_product (Ontology) int_pcrbait Dumps as: PCR_interactor <PCR_product> Bait
- PCR Target(s) - ?PCR_product (MultiOntology) int_pcrtarget Dumps as: PCR_interactor <PCR_product> Target
- Non-directional PCR(s) - ?PCR_product (MultiOntology) int_pcrnondir Dumps as: PCR_interactor <PCR_product> Non_directional
- Sequence Bait - ?Sequence (Free Text) int_sequencebait Dumps as: Sequence_interactor <Sequence> Bait
- Sequence Target(s) - ?Sequence (Free Text); separate multiple entries with pipes ('|') int_sequencetarget Dumps as: Sequence_interactor <Sequence> Target
- Non-directional Sequence(s) - ?Sequence (Free Text); separate multiple entries with pipes ('|') int_sequencenondir Dumps as: Sequence_interactor <Sequence> Non_directional
- Bait overlapping CDS - ?CDS (Free Text) int_cdsbait Dumps as: Interactor_overlapping_CDS <CDS> Bait
- Target overlapping CDS(s) - ?CDS (Free Text); separate multiple entries with pipes ('|') int_cdstarget Dumps as: Interactor_overlapping_CDS <CDS> Target
- Non-directional overlapping CDS(s) - ?CDS (Free Text); separate multiple entries with pipes ('|') int_cdsnondir Dumps as: Interactor_overlapping_CDS <CDS> Non_directional
- Bait overlapping protein - ?Protein (Free Text) int_proteinbait Dumps as: Interactor_overlapping_protein <Protein> Bait
- Target overlapping protein(s) - ?Protein (Free Text); separate multiple entries with pipes ('|') int_proteintarget Dumps as: Interactor_overlapping_protein <Protein> Target
- Non-directional overlapping protein(s) - ?Protein (Free Text); separate multiple entries with pipes ('|') int_proteinnondir Dumps as: Interactor_overlapping_protein <Protein> Non_directional
- Bait overlapping gene - ?Gene (Ontology) int_genebait Dumps as: Interactor_overlapping_gene <Gene> Bait
- Target overlapping gene(s) - ?Gene (MultiOntology) int_genetarget Dumps as: Interactor_overlapping_gene <Gene> Target
- Antibody - ?Antibody (MultiOntology) int_antibody Dumps as: Interactor_overlapping_gene <Mapped Gene> Antibody <Antibody> AND Antibody (on new line)
- When mapping antibodies to genes, compare antibody-affiliated genes with those in the Non-directional Gene(s), Effector Gene(s), Affected Gene(s), Bait Overlapping Gene and Target Overlapping Gene fields
- For Antibodies that don't map to a gene in the interaction, Dump as: Unaffiliated_antibody <Antibody>
- Antibody remark - Big Text int_antibodyremark Dumps as: Antibody_remark <Big_Text>
TAB3
- Non-directional Gene(s) - ?Gene (MultiOntology) int_genenondir Dumps as: Interactor_overlapping_gene <Gene> Non_directional
- Effector Gene(s) - ?Gene (MultiOntology) int_geneone Dumps as: Interactor_overlapping_gene <Gene> Effector
- Affected Gene(s) - ?Gene (MultiOntology) int_genetwo Dumps as: Interactor_overlapping_gene <Gene> Affected
- Non-directional Variation(s) - ?Variation (MultiOntology) int_variationnondir
- Genes for this field need to be mapped to a gene at the dump stage; Genes that map to the variation will be dumped as the "Interactor_overlapping_gene" as follows:
- Dumps as: (Line 1) Interactor_overlapping_gene <Mapped Gene> Variation <Variation>
- Dumps as: (Line 2) Interactor_overlapping_gene <Mapped Gene> Non_directional
- For Variations that don't map to a gene, Dump as: Unaffiliated_variation <Variation>
- Effector Variation(s) - ?Variation (MultiOntology) int_variationone
- Genes for this field need to be mapped to a gene at the dump stage; Genes that map to the variation will be dumped as the "Interactor_overlapping_gene" as follows:
- Dumps as: (Line 1) Interactor_overlapping_gene <Mapped Gene> Variation <Variation>
- Dumps as: (Line 2) Interactor_overlapping_gene <Mapped Gene> Effector
- For Variations that don't map to a gene, Dump as: Unaffiliated_variation <Variation>
- Affected Variation(s) - ?Variation (MultiOntology) int_variationtwo
- Genes for this field need to be mapped to a gene at the dump stage; Genes that map to the variation will be dumped as the "Interactor_overlapping_gene" as follows:
- Dumps as: (Line 1) Interactor_overlapping_gene <Mapped Gene> Variation <Variation>
- Dumps as: (Line 2) Interactor_overlapping_gene <Mapped Gene> Affected
- For Variations that don't map to a gene in the interaction, Dump as: Unaffiliated_variation <Variation>
- Intragenic Effector Variation(s) - ?Variation (MultiOntology) int_intravariationone
- Genes for this field need to be mapped to a gene at the dump stage; Genes that map to the variation will be dumped as the "Interactor_overlapping_gene" as follows:
- Dumps as: Interactor_overlapping_gene <Mapped Gene> Intragenic_effector_variation <Variation>
- For Variations that don't map to a gene in the interaction, Dump as: Unaffiliated_variation <Variation>
- Intragenic Affected Variation(s) - ?Variation (MultiOntology) int_intravariationtwo
- Genes for this field need to be mapped to a gene at the dump stage; Genes that map to the variation will be dumped as the "Interactor_overlapping_gene" as follows:
- Dumps as: Interactor_overlapping_gene <Mapped Gene> Intragenic_affected_variation <Variation>
- For Variations that don't map to a gene in the interaction, Dump as: Unaffiliated_variation <Variation>
TAB4
- Deviation from expectation - Big text int_deviation Dumps as: Deviation_from_expectation <Big_Text>
- Neutrality function - (Dropdown) int_neutralityfxn options are:
- Multiplicative Dumps as: Multiplicative
- Additive Dumps as: Additive
- Minimal Dumps as: Minimal
- Non-directional Rearrangement(s) - ?Rearrangement (MultiOntology) int_rearrnondir Dumps as: Rearrangement <Rearrangement> Non_directional
- Effector Rearrangement(s) - ?Rearrangement (MultiOntology) int_rearrone Dumps as: Rearrangement <Rearrangement> Effector
- Affected Rearrangement(s) - ?Rearrangement (MultiOntology) int_rearrtwo Dumps as: Rearrangement <Rearrangement> Affected
- Effector Other Type - (Dropdown) int_otheronetype options are: Chemical or Transgene, int_otheronetype Dumps as (see next line)
- Effector Other - ?Text int_otherone Dumps as: Remark "Effector <Effector Other Type>: <Text>"
- Affected Other Type - (Dropdown) int_othertwotype options are: Chemical or Transgene, int_othertwotype Dumps as (see next line)
- Affected Other - ?Text int_othertwo Dumps as: Remark "Affected <Affected Other Type>: <Text>"
- RNAi - (MultiOntology) int_rnai Dumps as: Interaction_RNAi <RNAi>
- Large scale RNAi - Free Text; separate multiple entries with pipes ('|') int_lsrnai (all large scale RNAi that doesn't match ontology) Dumps as: Interaction_RNAi <RNAi>
- Interaction phenotype(s) - ?Phenotype (MultiOntology) int_phenotype Dumps as: Interaction_phenotype <Phenotype>
- Expression pattern(s) - ?Expr_pattern (MultiOntology) int_exprpattern Dumps as: Interactor_overlapping_gene <Mapped Gene> Expr_pattern <Expr_pattern>
- When mapping Expression patterns to genes, compare Expr-affiliated genes with those in the Non-directional Gene(s), Effector Gene(s), Affected Gene(s), Bait Overlapping Gene and Target Overlapping Gene fields
- For Expression patterns that don't map to a gene in the interaction, Dump as: Unaffiliated_expr_pattern <Expr_pattern>
- Transgene(s) - ?Transgene (MultiOntology) int_transgene Dumps as: Interactor_overlapping_gene <Mapped Gene> Transgene <Transgene> AND Transgene (on new line)
- When mapping transgenes to genes, compare transgene-affiliated genes (from the Driven_by_gene, Gene, and 3'UTR fields) with those in the Non-directional Gene(s), Effector Gene(s), Affected Gene(s), Bait Overlapping Gene and Target Overlapping Gene fields
- For Transgenes that don't map to a gene in the interaction, Dump as: Unaffiliated_transgene <Transgene>
TAB5
- Person - ?Person int_person Dumps as: Remark <Remark_text> Person_evidence <Person>
- If there is no Remark entry, dumps as: Remark "See Person Evidence" Person_evidence <Person>
- Confidence description - Text int_confidence Dumps as: Description <Text>
- P-value - Text (Float) int_pvalue Dumps as: P_value FLOAT
- Log-likelihood score - Text (Float) int_loglikelihood Dumps as: Log_likelihood_score FLOAT
- High_throughput - (Toggle) int_throughput:
- If ON, dumps as: High_throughput
- If OFF (default), dumps as: Low_throughput
- Sentence ID - (Ontology) sentence shows in term info; int_sentid Dumps as: N/A
- False Positive - toggle, will not give an id or no dump if the sentence is false positive, containing no interaction info; int_falsepositive Dumps as: N/A
To go live on tazendra
To create new interaction tables on tazendra : /home/postgres/work/pgpopulation/interaction/20120527_OA_newModel/create_datatype_tables.pl
Backup relevant tables : /home/postgres/work/pgpopulation/interaction/20120527_OA_newModel/backupTable.pl
To transfer data from old interaction model tables to new interaction model tables and table formats : /home/postgres/work/pgpopulation/interaction/20120527_OA_newModel/transfer_int_data.pl
Instructions on How to Use the New OA
The new Interaction OA is intended to be used by curators for curating physical, predicted and genetic interactions. This is an overview of the key points to keep in mind while curating with the Interaction OA.
1) TAB 1 is for general information, TAB 2 is for physical interactions, TAB 3 & TAB 4 are for genetic and predicted interactions, and TAB5 is for detailed (and rarely used) information
TAB 1
2) Interaction IDs are generated automatically when the "New" button is clicked; new Postgres IDs (PGIDs) are generated automatically as well. If you would like to duplicate objects (because you are generating several similar interaction objects) and assign new IDs to them, select the interaction to duplicate and click the "Duplicate" button; this will generate a new PGID and OA row, but carry the same Interaction ID over from the duplicated interaction. If the new row should be a distinct interaction, delete the interaction ID from the Interaction ID field, and a cron job will assign an Interaction ID to that row the following evening. Note that if you leave any rows without an Interaction ID overnight, they will be each assigned a unique (and new) Interaction ID.
3) Entering Database information: Database information is typically provided with three pieces of information: the Database, the Database field name, and the Database Accession number for the interaction in question. These must be each entered surrounded by double quotes (") and separated with spaces. So for example:
"Database" "Database Field" "Accession Number"
If mulitiple database references are to be made, split on pipes ("|") like this:
"Database 1" "Database Field 1" "Accession Number 1" | "Database 2" "Database Field 2" "Accession Number 2"
In the (unlikely) event that any of the field entries have double quotes (") in the name itself, then the 'inner' double quotes will need to be escaped with a backslash ("\") like this:
"Database \"Supreme!!!!\"" "Database \"Field\"" "Accession \"Number\""
so that it will be read in ACEDB as:
Database "Supreme!!!!" Database "Field" Accession "Number"
4) The Person field is for Person evidence, when no other reference (such as a WormBase paper) is supplied as a reference. This will be dumped as a hash/supplement to the "Remark" entry, so it is advantageous to include any pertinent information there.
TAB 2
5) The "Library screens and Times found" field is for documenting screening/testing libraries that were used to identify a physical interaction. For example, a cDNA library may be used in a Yeast Two Hybrid screen to identify protein interaction partners with a "Bait" protein. Sometimes (but not always) authors might report the number of times a particular interaction was identified using a particular library. If not, enter the name of the library with double quotes (") like so:
"cDNA"
if multiple libraries (but no numbers), split on pipes like this:
"cDNA" | "ORFeome"
If a single libary, with a number (for number of times found):
"AD-TF mini-library" 5
and if multiple libraries, with numbers:
"AD-TF mini-library" 5 | "AD-wrmcDNA library" 1
As with the Database field entries, if (for some reason) the name of the library has double quotes (") in the name itself, the double quotes will need to be escaped like this:
"AD-TF \"Mini\" library" 5
so that the library name eventually reads like this:
AD-TF "Mini" library
6) Sequence fields: "Sequence Bait", "Sequence Target(s)", and "Non-directional Sequence(s)"
To enter a single sequence object, type in the sequence object name, no quotes:
CK583862
For multiple sequence objects, split on pipes ("|"):
CK583862 | CK583870
The same rules apply for Protein and CDS objects.
7) The Non-directional fields; For each type of interaction object, there is a "Non-directional" field, allowing a curator to enter all interactors of that type for a Non-directional type of interaction. Note that the "Non-directional Genes" field (which could apply to physical, genetic, and predicted interactions) lies in TAB 3.
8) For Directional interactions, there are "Bait" and "Target" fields for physical interactions (TAB 2) and "Effector" and "Affected" fields for genetic interactions (TAB 3).
TAB 3
9) Effector Variation(s), Affected Variation(s), and Non-directional Variation(s) fields are for variations that implicate an affiliated gene as an effector or affected interactor in a directional genetic interaction or non-directional interactors in a non-directional interaction. These fields can be populated without the need to populate the respective gene in the relevant gene field, as the dumping script will make the appropriate associations.
10) The Intragenic Effector Variation(s) and Intragenic Affected Variation(s) fields are reserved for genetic interactions between two variations within the same gene, for example intragenic suppression events. The lack of a "Non-directional" field assumes that all of these interactions will be directional.
TAB 4
11) Deviation from expectation is a free-text field where, at the curator's discretion, a curator can describe why a genetic interaction is an interaction, i.e. why it is an unexpected result warranting an interaction. This could be as simple as "The life spans were synergistic" or "Neither mutation alone exhibited a phenotype".
12) Neutrality function is closely related to the "Deviation from expectation" field, where the curator can decide, if applicable, which "Neutrality function" applies to this genetic interaction. The choices in this drop down field are "Multiplicative", "Additive", and "Minimal". "Multiplicative" means that the authors expected to see a quantifiable phenotype in the double mutant that was the mathematical product of the quantified phenotypes of the individual mutations. For example, one mutant extends life span by 20%, and another extends it by 30%. With a multiplicative neutrality function, we would expect the double mutant to have (1.2 * 1.3 = 1.56) about 56% extended life span. Alternatively, in the "Additive" neutrality function, the authors might expect the sum of the effects (A + B - 1) or (1.2 + 1.3 - 1 = 1.5) or a 50% extended life span. The "Minimal" neutrality function assumes that the double mutant will be as severe as the most severe single mutant, in this case (1.3) or 30% extended. Therefore, any life span extension beyond 30%, for example, in the double mutant would be considered "surprising" and therefore, an interaction.
13) The RNAi fields: There are two fields for references to RNAi objects: "RNAi" and "Large scale RNAi". The reason for two fields is that one field ("RNAi") is an ontology field reading off of RNAi experiments that live in the RNAi OA and Postgres. As RNAi experiments from papers containing 2,000 or more RNAi experiments (WBPaper00029258 for example) were excluded from the RNAi OA for performance reasons, any RNAi experiments from such papers will not be recognized by the "RNAi" ontology field, and therefore must be entered as free-text in the "Large scale RNAi" field.
14) The Transgene(s) field is for transgenes involved in the interaction, regardless of whether it is related to an "Effector" gene or "Affected" gene or the interaction is Non-directional. The dumping script will automatically associate the transgene with the appropriate interacting gene and dump in .ACE format accordingly.
15) Effector/Affected Other Type and Effector/Affected Other fields: these fields allow for the curation of Chemicals, Transgenes, or other entities that don't exists as proper WormBase/ACEDB objects, for example transgenes that only express human proteins. The "Other Type" fields allow for the selection of "Chemical" or "Transgene", the identity of which would go into the "Other" fields. This, ideally, will get phased out as chemicals are generated in the Molecule OA and transgenes in the Transgene OA, thereby allowing them to be entered into ontology-based "Transgene" or "Molecule" fields.
TAB 5
16) Confidence description: This field will capture free-text descriptions of the confidence the authors suggest they have for this interaction. In the Yeast Two Hybrid experiments, for example, the interaction may be described as "Interactome Core 1", "Interactome Core 2", or "Interactome Noncore" referring to the varying degrees of confidence for those interactions.
17) The P-value and Log-likelihood score fields are mostly to capture confidence values for predicted interactions that have been reported.
18) High_throughput toggle field is intended to capture whether or not the interaction was observed as one of several (50 - 1000s) interactions and thus should be interpreted with caution, or at least acknowledged as from a large scale experiment. The default is OFF and indicates that the experiment is low throughput.
19) The Sentence ID and False Positve fields are exclusively for Textpresso sentence-based curation
Nightly Cron Job to Assign New Interaction IDs
Every night at 4am a cron job script will run to assign new Interaction IDs to any row/PGID in the Interaction OA that does not already have an Interaction ID and that meets a few criteria. The script is located on Tazendra here:
/home/acedb/xiaodong/assigning_interaction_ids/assign_interaction_ids.pl
and the criteria for getting a new Interaction ID are as follows:
1) The curator of the interaction object/PGID is NOT Arun
2) The interaction object/PGID does NOT already have an Interaction ID
3) The interaction object/PGID is NOT flagged as False Positive
Any interactions/PGIDs that meet these three criteria will be assigned a new Interaction ID by the cron job.
Interaction OA .ACE Dumper
The script for the interaction OA dumper is located on Tazendra at:
/home/postgres/work/citace_upload/interaction/use_package.pl*
Error Checks During Dump Process
The following is a list of checks that the .ACE dumper script will perform on all interactions being dumped out of the OA to make sure that the data is consistent and doesn't have any nonsensical information:
Fatal Errors (Interactions will not get dumped)
1) If there are fewer than two interactors in an interaction, the dumper script will generate an error message that is printed to the ERROR output file and the object will not get dumped. This is determined by checking that:
a) There is at least one "Bait" entry and one "Target" entry OR
b) There is at least one "Effector" and one "Affected" entry OR
c) There is at least two "Non-directional" entries OR
d) There is at least one "Intragenic Effector Variation" and at least one "Intragenic Affected Variation" entry
If none of these conditions hold true, then an error message will be printed in tab-delimited format like this:
PGID <TAB> Dump_status <TAB> Curator ID <TAB> Explanation
so, for example:
12345 nodump WBPerson1234 There are not two interactors
2) If there is no reference (Paper or Person) then the object will not get dumped and an error message is printed:
PGID <TAB> Dump_status <TAB> Curator ID <TAB> Explanation
so, for example:
12345 nodump WBPerson1234 There is no reference, neither paper nor person
3) If there are incompatible interactor types, the interaction object will not get dumped. This means that the object will not get dumped if the following conditions are not met:
a) If there is a "Non-directional" entry, there are no "Effector", "Affected", "Bait", or "Target" entries AND
b) If there is an "Effector" entry, there is at least one "Affected" entry AND there are no "Non-directional", "Bait", or "Target" entries AND
c) If there is an "Affected" entry, there is at least one "Effector" entry AND there are no "Non-directional", "Bait", or "Target" entries AND
d) If there is a "Bait" entry, there is at least one "Target" entry AND there are no "Non-directional", "Effector", or "Affected" entries AND
e) If there is a "Target" entry, there is at least one "Bait" entry AND there are no "Non-directional", "Effector", or "Affected" entries
If these conditions are not met, the object will not get dumped and an error message will print to the ERROR output file like this:
PGID <TAB> Dump_status <TAB> Curator ID <TAB> Explanation
so, for example:
12345 nodump WBPerson1234 has nondiretional + bait 12345 nodump WBPerson1234 has nondiretional + target 12345 nodump WBPerson1234 has nondiretional + effected 12345 nodump WBPerson1234 has nondiretional + effector 12345 nodump WBPerson1234 has effector but no effected 12345 nodump WBPerson1234 has effector + bait 12345 nodump WBPerson1234 has effector + target 12345 nodump WBPerson1234 has effected + bait 12345 nodump WBPerson1234 has effected + target 12345 nodump WBPerson1234 has bait but no target
4) If there is no Interaction ID, the object will not get dumped and an error message will print to the ERROR output file like this:
PGID <TAB> Dump_status <TAB> Curator ID <TAB> Explanation
so, for example:
12345 nodump WBPerson1234 There is no Interaction ID
The script will determine this by 1) generating a list of all PGIDs from the Interaction OA, 2) Removing all PGIDs where Arun is the curator, and then 3) looking for any PGIDs for which there is no Interaction ID. As there is a cronjob to add Interaction IDs to any PGIDs (Postgres rows) that are missing IDs, this problem should be rare (unless objects have been added that day (before the next cronjob) without Interaction IDs).
5) If there is no Interaction Type, the object will not get dumped and an error message will print to the ERROR output file like this:
PGID <TAB> Dump_status <TAB> Curator ID <TAB> Explanation
so, for example:
12345 nodump WBPerson1234 There is no Interaction Type
6) If there is an Interaction object that exists on multiple Postgres lines/rows (i.e. the same Interaction ID with multiple PGIDs), the object will not get dumped and an error message will print to the ERROR output file like this:
PGID <TAB> Dump_status <TAB> Curator ID <TAB> Explanation
so, for example:
12345 nodump WBPerson1234 WBInteraction000123456 exists across multiple lines
Non-Fatal Errors (Interactions will get dumped, but error message will get printed)
1) If there is a Variation, Expression pattern, Transgene, or Antibody that cannot be matched to a gene interactor, the object will be identified as "Unaffiliated" in the .ACE file and an error message will print to the ERROR output file like this:
PGID <TAB> Dump_status <TAB> Curator ID <TAB> Interaction ID <TAB> Unaffiliated Object <TAB> Object Name
so, for example:
12345 lineonly WBPerson1234 WBInteraction000123456 Unaffiliated_variation WBVar00600763 12345 lineonly WBPerson1234 WBInteraction000123456 Unaffiliated_transgene kyEx456 12345 lineonly WBPerson1234 WBInteraction000123456 Unaffiliated_antibody [cgc2826]:hlh-2 12345 lineonly WBPerson1234 WBInteraction000123456 Unaffiliated_expr_pattern Expr1234
2) If there is an inconsistency between the directionality of the Interaction Type and the Interactors, the interaction object will get dumped to the .ACE file, but an error message will print to the ERROR output file like this:
PGID <TAB> Dump_status <TAB> Curator ID <TAB> Explanation
so, for example:
12345 flagonly WBPerson1234 has diretional type Enhancement + nondirectional data 12345 flagonly WBPerson1234 has diretional type Epistasis + nondirectional data 12345 flagonly WBPerson1234 has diretional type Suppression + nondirectional data 12345 flagonly WBPerson1234 has nondiretional type Mutual_enhancement + effected data 12345 flagonly WBPerson1234 has nondiretional type Mutual_enhancement + effector data 12345 flagonly WBPerson1234 has nondiretional type Mutual_suppression + effector data 12345 flagonly WBPerson1234 has nondiretional type No_interaction + effected data 12345 flagonly WBPerson1234 has nondiretional type Synthetic + effector data
3) If no curator is listed for the interaction, the interaction object will get dumped, but an error message will print to the ERROR output file like this:
PGID <TAB> Dump_status <TAB> Curator ID <TAB> Explanation
so, for example:
12345 flagonly no curator has no curator
Handling Dead Genes During Dump Process
The dumper script will now (as of May, 2013) run an automatic check for dead genes in any gene field. Any genes that are considered dead that are referenced in an Interaction object in the OA will be handled in the following manner:
1) If there is a replacement for the gene (i.e. the gene has merged into another gene), the dead gene will be dumped into a "Historical_gene" field in the .ACE file, the replacement gene will fill the original gene field. A comment will be added to the Historical_gene field via the #Evidence hash. The original gene field (now with the updated gene reference) will be printed with an "Inferred_automatically" tag after the gene. So, for example, if WBGene00001234 is now a dead gene that has been merged into WBGene00002345:
Gene "WBGene00001234"
becomes
Gene "WBGene00002345" Inferred_automatically Historical_gene "WBGene00001234" Remark "Note: This object originally referred to WBGene00001234. WBGene00001234 is now considered dead and has been merged into WBGene00002345. WBGene00002345 has replaced WBGene00001234 accordingly."
Also, since Antibodies, Transgenes, Expression patterns, Variations are mapped to an interactor where possible (or else they are dumped as "Unaffiliated"), this mapping will now occur to only the newest genes that the interactor refers to.
2) If there is no replacement for the gene, we would dump the following:
Historical_gene "WBGene00001234" Remark "Note: This object originally referred to an interacting gene (WBGene00001234) that is now considered dead. Please interpret with discretion."
and lastly,
3) If the gene has undergone a split, such genes will be printed out in the error output file of the dumping script for a curator to go back and manually change according to best judgement.