Contributing Allele Phenotype Connections
The following documentation explains the use and backend for the Allele-Phenotype form found here:
http://tazendra.caltech.edu/~azurebrd/cgi-bin/forms/allele_phenotype.cgi
Contents
User Guide
Finding the Allele-Phenotype form
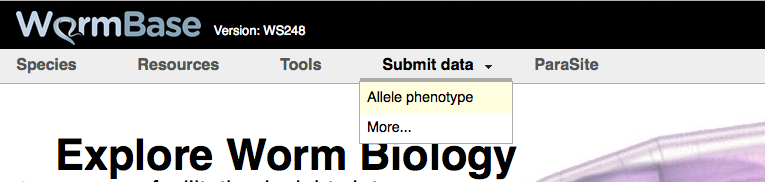
There are two main ways to reach the Allele-Phenotype form from the WormBase homepage. First, and most directly, you can click on the "Allele-Phenotype" option under the "Submit data" menu header at the top of the WormBase homepage:
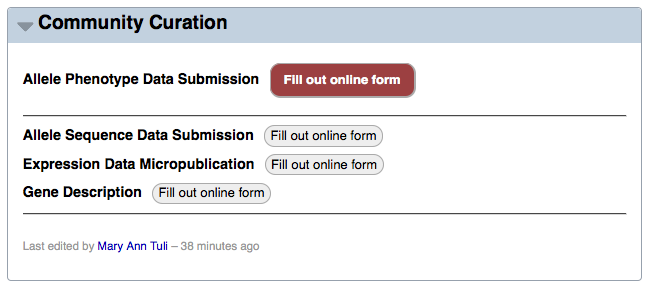
Second, the data submission summary page (accessible by clicking on the "Submit data" menu header at the top of the WormBase homepage) has a link to the form via the red "Fill out online form" button to the right of "Allele Phenotype Data Submission":
Minimum required information for data submission
The form minimally requires a submitter's name and e-mail address, a publication's PubMed ID, an allele name, and a phenotype (observed or NOT observed).
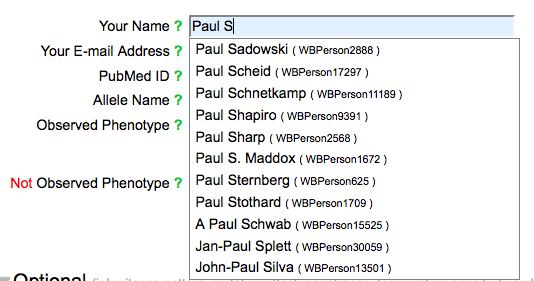
Your Name & E-mail Address
The name field has an autocomplete functionality, requiring the selection of a name from the list of registered WormBase Persons.
If your name is not registered with WormBase, click on the green question mark to the left of the name field to open up the help text in the term information box in the upper right-hand corner of the screen and click on the "Person Update Form" link to register. If WormBase has your e-mail address on file, it will automatically be filled into the e-mail field, otherwise you may add (or correct) your e-mail address manually.
PubMed ID
Once a name has been selected, a notice will appear to the right of the name field with a link to a summary of your publications and their associated PubMed IDs:
The publication summary page consists of two tables: the top table lists papers for which WormBase data flagging pipelines have predicted the presence of allele-phenotype data and the bottom table lists papers that have not been flagged for allele-phenotype data.
- Image here
The top table lists the curation status of papers, indicating that the paper either "Needs curation" (i.e. there are no allele-phenotype annotations for this paper), has "Curation in progress" (i.e. there is at least one annotation from a community curator using this form) or is "WormBase curated" (i.e. this paper has been curated by WormBase curators). Clicking on the "Needs curation" link will direct you to back to the Allele-Phenotype form with your name, e-mail address (if already provided), and PubMed ID of that paper pre-populated into the appopriate fields of the form:
- Image here