Difference between revisions of "WormBase Genomes"
(finished adding strains) |
m |
||
| Line 43: | Line 43: | ||
! Origin | ! Origin | ||
! Comments | ! Comments | ||
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 51: | Line 52: | ||
| Yes | | Yes | ||
| WashU | | WashU | ||
| − | | [Sept 2010] New assembly from Erich Haag being worked on. [Feb 2011] updated in WS224 | + | | First added in WS132<br>[Sept 2010] New assembly from Erich Haag being worked on.<br>[Feb 2011] updated in WS224 |
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 61: | Line 63: | ||
| WashU | | WashU | ||
| released in WS226. '''WARNING''' the genome sequence contains contaminations | | released in WS226. '''WARNING''' the genome sequence contains contaminations | ||
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 70: | Line 73: | ||
| WashU | | WashU | ||
| released in WS226. | | released in WS226. | ||
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 79: | Line 83: | ||
| WashU | | WashU | ||
| | | | ||
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 88: | Line 93: | ||
| WashU | | WashU | ||
| genome and deNovo gene set released in WS226.<br>Replaced by RNAseq based gene set in WS227. | | genome and deNovo gene set released in WS226.<br>Replaced by RNAseq based gene set in WS227. | ||
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 97: | Line 103: | ||
| WashU | | WashU | ||
| [Jan 2011] The current assembly contains quite a bit of heterozygosity.<br>'''Warning''' WS223-WS226 genome assembly is not in sync with the annotation files or INSDC. Please use WS227+ | | [Jan 2011] The current assembly contains quite a bit of heterozygosity.<br>'''Warning''' WS223-WS226 genome assembly is not in sync with the annotation files or INSDC. Please use WS227+ | ||
| + | |||
| + | |||
|-bgcolor="#FFFF33" | |-bgcolor="#FFFF33" | ||
| V | | V | ||
| Line 105: | Line 113: | ||
| Yes | | Yes | ||
| WashU/Sanger | | WashU/Sanger | ||
| − | | | + | | First added in WS1 |
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 115: | Line 124: | ||
| WashU | | WashU | ||
| released in WS226.<br>'''WARNING''' the genome sequence contains contaminations | | released in WS226.<br>'''WARNING''' the genome sequence contains contaminations | ||
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 124: | Line 134: | ||
| WashU | | WashU | ||
| [Jan 2011] New/improved assembly is being worked on at WashU.<br>[Oct 2011] new assembly in WS227 | | [Jan 2011] New/improved assembly is being worked on at WashU.<br>[Oct 2011] new assembly in WS227 | ||
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 133: | Line 144: | ||
| CalTech | | CalTech | ||
| Added to WormBase in release WS218.<br>[Jan 2011] This species now has an official name of '''C. angaria''' | | Added to WormBase in release WS218.<br>[Jan 2011] This species now has an official name of '''C. angaria''' | ||
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 142: | Line 154: | ||
| Sanger | | Sanger | ||
| Added to WormBase in release WS209. | | Added to WormBase in release WS209. | ||
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 151: | Line 164: | ||
| WashU | | WashU | ||
| [Jan 2011] Submitted to GenBank - Accession: EF043402<br>[Sep 2011] Gene set and Annotations are being worked on.<br>[Nov 2011] on WormBase as of WS229 | | [Jan 2011] Submitted to GenBank - Accession: EF043402<br>[Sep 2011] Gene set and Annotations are being worked on.<br>[Nov 2011] on WormBase as of WS229 | ||
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 160: | Line 174: | ||
| WashU/MPI | | WashU/MPI | ||
| [16 December 2010] Updated to the newest assembly and geneset in WS221 | | [16 December 2010] Updated to the newest assembly and geneset in WS221 | ||
| + | |||
|-bgcolor="#FFFF99" | |-bgcolor="#FFFF99" | ||
| V | | V | ||
| Line 169: | Line 184: | ||
| [http://www.langebio.cinvestav.mx/ LANGEBIO] / CalTech | | [http://www.langebio.cinvestav.mx/ LANGEBIO] / CalTech | ||
| [May 2011] Being assembled. | | [May 2011] Being assembled. | ||
| + | |||
|-bgcolor="#99FF99" | |-bgcolor="#99FF99" | ||
| IV | | IV | ||
| Line 178: | Line 194: | ||
| INRA | | INRA | ||
| Added to WormBase in release WS205.<br>Genes are not yet available. | | Added to WormBase in release WS205.<br>Genes are not yet available. | ||
| + | |||
|-bgcolor="#99FF99" | |-bgcolor="#99FF99" | ||
| IV | | IV | ||
| Line 187: | Line 204: | ||
| NCSU hapla.org | | NCSU hapla.org | ||
| Added to WormBase in release WS204. | | Added to WormBase in release WS204. | ||
| + | |||
|-bgcolor="#99FF99" | |-bgcolor="#99FF99" | ||
| IV | | IV | ||
| Line 196: | Line 214: | ||
| Sanger | | Sanger | ||
| [Jan 2011] draft assembly in GenBank, released in WS226 | | [Jan 2011] draft assembly in GenBank, released in WS226 | ||
| + | |||
|-bgcolor="#99FF99" | |-bgcolor="#99FF99" | ||
| IV | | IV | ||
| Line 205: | Line 224: | ||
| Sanger | | Sanger | ||
| [Sept 2011] published in PLOS Pathogens<br>[Nov 2011] added to WormBase in WS229 | | [Sept 2011] published in PLOS Pathogens<br>[Nov 2011] added to WormBase in WS229 | ||
| + | |||
|-bgcolor="#FF9900" | |-bgcolor="#FF9900" | ||
| III | | III | ||
| Line 214: | Line 234: | ||
| Davis | | Davis | ||
| [Oct 2011] integrated the Davis genome without a reference gene set.<br>[Nov 2011] added a reference gene set | | [Oct 2011] integrated the Davis genome without a reference gene set.<br>[Nov 2011] added a reference gene set | ||
| + | |||
|-bgcolor="#FF9900" | |-bgcolor="#FF9900" | ||
| III | | III | ||
| Line 223: | Line 244: | ||
| TIGR -> WashU/Sanger | | TIGR -> WashU/Sanger | ||
| [Sept 2010] Currently using the old TIGR assembly. Waiting for WashU (did assembly) and Sanger (did gene models) to publish, then we will use the new assembly.<br>[Dec 2010] merged Augustus gene predictions from Erich Schwarz into WS216 | | [Sept 2010] Currently using the old TIGR assembly. Waiting for WashU (did assembly) and Sanger (did gene models) to publish, then we will use the new assembly.<br>[Dec 2010] merged Augustus gene predictions from Erich Schwarz into WS216 | ||
| + | |||
|-bgcolor="#33FFFF" | |-bgcolor="#33FFFF" | ||
| I | | I | ||
| Line 248: | Line 270: | ||
! Origin | ! Origin | ||
! Comments | ! Comments | ||
| + | |||
|-bgcolor="#CC9999" | |-bgcolor="#CC9999" | ||
| V | | V | ||
| Line 257: | Line 280: | ||
| WashU | | WashU | ||
| [Sept 2010] being assembled | | [Sept 2010] being assembled | ||
| + | |||
|-bgcolor="#CC9999" | |-bgcolor="#CC9999" | ||
| V | | V | ||
| Line 266: | Line 290: | ||
| Mark Blaxter | | Mark Blaxter | ||
| status unclear | | status unclear | ||
| + | |||
|-bgcolor="#CC9999" | |-bgcolor="#CC9999" | ||
| | | | ||
| Line 275: | Line 300: | ||
| Sanger | | Sanger | ||
| | | | ||
| + | |||
|-bgcolor="#CC9999" | |-bgcolor="#CC9999" | ||
| | | | ||
| Line 284: | Line 310: | ||
| Sanger | | Sanger | ||
| | | | ||
| + | |||
|-bgcolor="#CC9999" | |-bgcolor="#CC9999" | ||
| | | | ||
| Line 293: | Line 320: | ||
| Sanger | | Sanger | ||
| | | | ||
| + | |||
|-bgcolor="#CC9999" | |-bgcolor="#CC9999" | ||
| | | | ||
| Line 302: | Line 330: | ||
| Sanger | | Sanger | ||
| | | | ||
| + | |||
|-bgcolor="#CC9999" | |-bgcolor="#CC9999" | ||
| | | | ||
| Line 311: | Line 340: | ||
| Sanger | | Sanger | ||
| | | | ||
| + | |||
|-bgcolor="#CC9999" | |-bgcolor="#CC9999" | ||
| | | | ||
Revision as of 12:30, 15 December 2011
Contents
WormBase Genomes
This is a record of the current and proposed set of genomes in WormBase.
We may, of course, alter our plans for which species to include as circumstances dictate and so the list of organisms which should be included should be treated as somewhat tentative.
Clade
The Major Clades of Blaxter et al 1998 ("Bclades"), as systematised by De Ley and Blaxter 2002-2004.
Tiers
The different genomes in WormBase are classified in various tiers which depend on the amount of curation effort we are able to put into maintaining them.
Tier I - All efforts are made to curate the gene structures and any other genetic or metabolic information. Only C. elegans is in this group.
Tier II - Efforts are made, where practical, to manually curate the gene structure and possibly some other genomic information. WormBase 'owns' the assembly in the ENA and GenBank so that new gene annotations can be submitted to the ENA/GenBank by WormBase.
Tier III - No curation by WormBase. We will set up the genome on WormBase with any gene structures that the authors of this genome have predicted.
Tier IV - No curation by WormBase. Only transcriptome information provided by the authors and no coherent genome.
Tier V - No curation by WormBase. Only genome information provided by the authors and no coherent transcriptome.
Genome
The Genome column in the table gives the assembly size and a link to the genome in WormBase, or the approximate size if it has not been assembled.
Gene
The Genes column in the table indicates whether gene structures have been added to WormBase.
The current genomes
| Clade | Species | NCBI Taxon | Tier | Genome | Genes | Origin | Comments |
|---|---|---|---|---|---|---|---|
| V | Caenorhabditis briggsae strain AF16 |
6238 | II | 108419768 bp | Yes | WashU | First added in WS132 [Sept 2010] New assembly from Erich Haag being worked on. [Feb 2011] updated in WS224 |
| V | Caenorhabditis species 9 strain JU1422 |
870437 | V | 204396809 bp | WashU | released in WS226. WARNING the genome sequence contains contaminations | |
| V | Caenorhabditis species 5 | ? | V | WashU | released in WS226. | ||
| V | Caenorhabditis remanei | 31234 | II | 145500347 bp | Yes | WashU | |
| V | Caenorhabditis species 11 strain JU1373 |
886184 | III | 79321433 bp | Yes | WashU | genome and deNovo gene set released in WS226. Replaced by RNAseq based gene set in WS227. |
| V | Caenorhabditis brenneri (species 4) |
135651 | II | 190421492 bp | Yes | WashU | [Jan 2011] The current assembly contains quite a bit of heterozygosity. Warning WS223-WS226 genome assembly is not in sync with the annotation files or INSDC. Please use WS227+
|
| V | Caenorhabditis elegans strain Bristol N2 |
6239 | I | 100272276 bp | Yes | WashU/Sanger | First added in WS1 |
| V | Caenorhabditis species 7 strain JU1286 |
870436 | V | WashU | released in WS226. WARNING the genome sequence contains contaminations | ||
| V | Caenorhabditis japonica strain DF5080 |
281687 | II | 166565019 bp | Yes | WashU | [Jan 2011] New/improved assembly is being worked on at WashU. [Oct 2011] new assembly in WS227 |
| V | Caenorhabditis angaria strain PS1010 (species 3) |
96668 | III | 79761545 bp | Yes | CalTech | Added to WormBase in release WS218. [Jan 2011] This species now has an official name of C. angaria |
| V | Haemonchus contortus | 6289 | III | 297975349 bp | Yes | Sanger | Added to WormBase in release WS209. |
| V | Heterorhabditis bacteriophora strain M31e |
37862 | V | 76974349 bp | WashU | [Jan 2011] Submitted to GenBank - Accession: EF043402 [Sep 2011] Gene set and Annotations are being worked on. [Nov 2011] on WormBase as of WS229 | |
| V | Pristionchus pacificus strain PS312 |
54126 | II | 172773083 bp | Yes | WashU/MPI | [16 December 2010] Updated to the newest assembly and geneset in WS221 |
| V | Steinernema carpocapsae | 34508 | III | 230 Mb | LANGEBIO / CalTech | [May 2011] Being assembled. | |
| IV | Meloidogyne incognita | 6306 | III | 82095019 bp | INRA | Added to WormBase in release WS205. Genes are not yet available. | |
| IV | Meloidogyne hapla | 6305 | III | 53017507 bp | Yes | NCSU hapla.org | Added to WormBase in release WS204. |
| IV | Strongyloides ratti natural isolate |
34506 | III | 52638471 bp | Sanger | [Jan 2011] draft assembly in GenBank, released in WS226 | |
| IV | Bursaphelenchus xylophilus strain Ka4C1 |
6326 | III | 74561461 bp | Yes | Sanger | [Sept 2011] published in PLOS Pathogens [Nov 2011] added to WormBase in WS229 |
| III | Ascaris suum natural isolate |
6253 | III | 272782664 bp | Yes | Davis | [Oct 2011] integrated the Davis genome without a reference gene set. [Nov 2011] added a reference gene set |
| III | Brugia malayi natural isolate |
6279 | III | 95814443 bp | Yes | TIGR -> WashU/Sanger | [Sept 2010] Currently using the old TIGR assembly. Waiting for WashU (did assembly) and Sanger (did gene models) to publish, then we will use the new assembly. [Dec 2010] merged Augustus gene predictions from Erich Schwarz into WS216 |
| I | Trichinella spiralis | 6334 | III | 56779425 bp | Yes | WashU | [Sept 2010] Being assembled. [Feb 2011] published in Nature. [Mar 2011] added to WormBase in WS225 |
Genomes coming soon
| Clade | Species | NCBI Taxon | Tier | Genome | Genes | Origin | Comments |
|---|---|---|---|---|---|---|---|
| V | Caenorhabditis drosophilae | 96641 | III | WashU | [Sept 2010] being assembled | ||
| V | Caenorhabditis elegans strain DR1035 |
6239 | III | 100 Mb | Mark Blaxter | status unclear | |
| Onchocerca volvulus | 6282 | III | Sanger | ||||
| Globodera pallida | 36090 | III | Sanger | ||||
| Nippostrongylus brasiliensis | 27835 | III | Sanger | ||||
| Strongyloides ransomi | 553534 | III | Sanger | ||||
| Teladorsagia circumcincta | 45464 | III | Sanger | ||||
| Trichuris muris | 70415 | III | Sanger |
Phylogeny
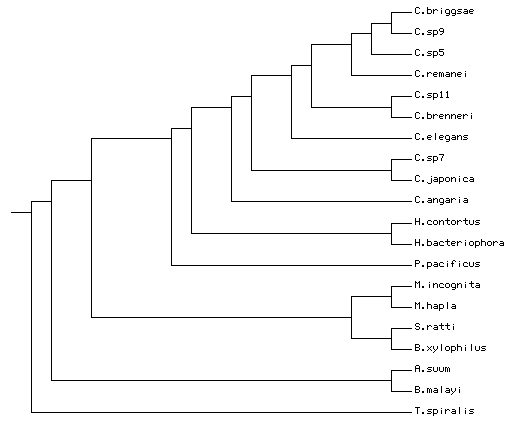
Given my understanding of the current phylogenetic literature (and based on personal communications with Karin Kiontke,David Fitch and Mark Blaxter), the correct guide tree would be:
((((((((((((C.briggsae,C.sp9),C.sp5),C.remanei),(C.sp11,C.brenneri)),C.elegans),(C.sp7,C.japonica)),C.angaria),(H.contortus,H.bacteriophora)),P.pacificus),((M.incognita,M.hapla), (S.ratti,B.xylophilus))),(A.suum,B.malayi)),T.spiralis);