TransgeneOme import
WormBase will import expression data (Images, constructs, and annotations) from the TransgeneOme project -Sarov et al., Cell, 2012. WBPaper00041419. The original data is always present on the Max Planck Institute (MPI) website. WormBase will fetch the data periodically (daily? Release-basis?) to have the collection up-to-date. WormBase does not provide means of annotating the patterns, hence community curation will happen on the TransgeneOme MPI site.
Contents
Data we want to import
What we want to have for each gene that has <Explore localization> data, is a table with Gene Strain image_name life_stage Annotations_by Anatomy_annotations Subcellular_annotations Construct
Ideally a tab delimited file with pipe separated entries for multiple entries, for example: WBGene00000095\tOP124\tL4larva\tMihail Sarov\tintestine|gonadal primordium|head\tnucleus\t7842083679062579 H01
JSON format is also good
Image format
We would like to have the images in JPG format
Image naming convention
- Daniela: Do the images have permanent IDs? If you replace the image with a higher resolution version, does that get a new ID that gets assigned to the gene while the previous image ID association gets removed?
- Stephan: The imaging works in our DB like this. We have imaging data. It has some metadata (created, creator, width, height) and a list of imaging data
channels (can be one of ZVI, DIC, GREEN, RED, BLUE, OTHER). The channel need to have same resolutions. Generally all entities in our DB get unique IDs. So if you replace a low resolution version of a DIC image in a imaging data (set), the imaging data ID stays the same, but the single imaging data ID changes.
- Daniela: How to see the file name of the image. For example 'strain-OP124-aha-1.png' for https://transgeneome.mpi-cbg.de/transgeneomics/user/downloadWidOverlay.html?id=31873093
- Stephan: The filenames for the overlays are not the real ones. These are generated on the fly. A real imaging data channel filename looks like
that: imagingdata-59891682-DIC-1.tiff So we store the original uploaded file renamed having the imaging data ID in it. The ID for that image would be 59891683.
Transgeneome DB API
The goal is an API for the Transgeneome DB in order to provide information about
- constructs
- strains
- images
- image annotations.
Export format will be JSON by default. For images it will be JPEG.
Things to consider:
- different species in TRG DB (request parameter)
* c.ele and d.mel
- genome (assembly versions)
- export image format, jpeg by default
- only provide public data
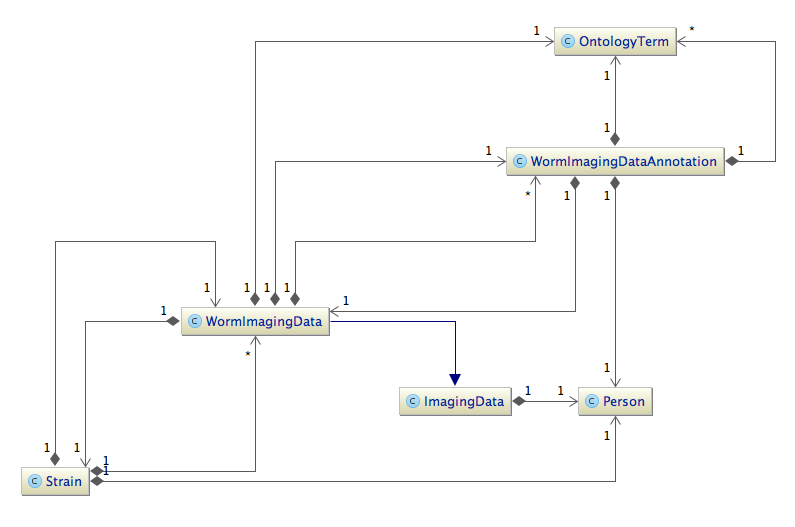
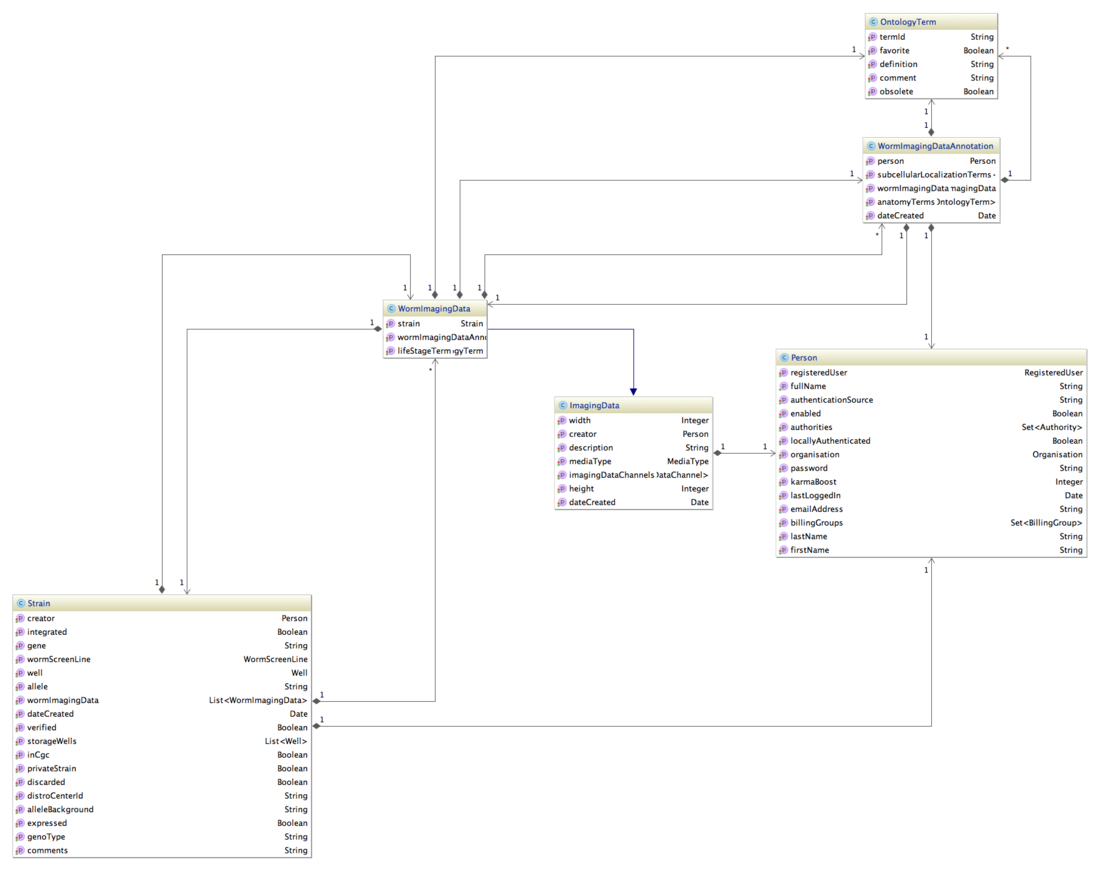
Transgeneome DB Data model
Here is a part of our class diagram without attributes. Excluded is the relation strain - well - feature - tag, which connects a strain to a gene.
Showing also the attributes helps to decode the json below immensely. Excluded is the relation strain - well - feature - tag, which connects a strain to a gene.
ImagingData
https://transgeneome.mpi-cbg.de/transgeneomics/api/imagingData.json
Example:
[
{
"lifeStageTerm": {
"termId": "WBls:0000057",
"name": "adult hermaphrodite",
"id": 7160
},
"strain": {
"well": {
"wellRow": "D",
"wellColumn": 6,
"selectedFeature": {
"tags": [
{
"field": "identifier",
"value": "WBGene00006375",
"id": 49490539
},
{
"field": "name",
"value": "syp-1",
"id": 49490541
},
{
"field": "id",
"value": "830756",
"id": 49490538
},
{
"field": "alias",
"value": "F26D2.2",
"id": 49490540
}
],
"name": null,
"id": 830756
},
"geneUnchangedFor": null,
"externalDbId": null,
"id": 13985414
},
"creator": {
"firstName": "Mihail",
"lastName": "Sarov",
"wormbaseID": "WBPerson3853",
"name": "sarov",
"id": 15
},
"dateCreated": "Mar 2, 2011",
"integrated": true,
"distroCenterId": null,
"allele": "ddIs226",
"alleleBackground": "unc-119(ed3)",
"genoType": null,
"url": null,
"name": "TH379",
"id": 32736214
},
"annotations": [
{
"dateCreated": "Jul 14, 2011",
"person": {
"firstName": "Mihail",
"lastName": "Sarov",
"wormbaseID": "WBPerson3853",
"name": "sarov",
"id": 15
},
"anatomyTerms": [
{
"termId": "WBbt:0005175",
"name": "gonad",
"id": 6191
}
],
"subcellularLocalizationTerms": [
{
"termId": "GO:0000795",
"name": "synaptonemal complex",
"id": 7677
}
],
"id": 46650021
}
],
"width": null,
"height": null,
"creator": {
"firstName": "TransgeneOmics Facility",
"lastName": "",
"wormbaseID": null,
"name": "schloissnig@mpi-cbg.de",
"id": 45762821
},
"dateCreated": "Mar 2, 2011",
"imagingDataChannels": [
{
"channelType": "GREEN",
"filename": "imagingdata-32736227-GREEN-1.jpg",
"id": 32736228
},
{
"channelType": "DIC",
"filename": "imagingdata-32736227-DIC-1.jpg",
"id": 32736229
}
],
"id": 32736227
},
{
"lifeStageTerm": {
"termId": "WBls:0000057",
"name": "adult hermaphrodite",
"id": 7160
},
"strain": {
"well": {
"wellRow": "D",
"wellColumn": 6,
"selectedFeature": {
"tags": [
{
"field": "identifier",
"value": "WBGene00006375",
"id": 49490539
},
{
"field": "name",
"value": "syp-1",
"id": 49490541
},
{
"field": "id",
"value": "830756",
"id": 49490538
},
{
"field": "alias",
"value": "F26D2.2",
"id": 49490540
}
],
"name": null,
"id": 830756
},
"geneUnchangedFor": null,
"externalDbId": null,
"id": 13985414
},
"creator": {
"firstName": "Mihail",
"lastName": "Sarov",
"wormbaseID": "WBPerson3853",
"name": "sarov",
"id": 15
},
"dateCreated": "Mar 2, 2011",
"integrated": true,
"distroCenterId": null,
"allele": "ddIs226",
"alleleBackground": "unc-119(ed3)",
"genoType": null,
"url": null,
"name": "TH379",
"id": 32736214
},
"annotations": [],
"width": null,
"height": null,
"creator": {
"firstName": "TransgeneOmics Facility",
"lastName": "",
"wormbaseID": null,
"name": "schloissnig@mpi-cbg.de",
"id": 45762821
},
"dateCreated": "Mar 2, 2011",
"imagingDataChannels": [
{
"channelType": "GREEN",
"filename": "imagingdata-32736230-GREEN-1.jpg",
"id": 32736231
},
{
"channelType": "DIC",
"filename": "imagingdata-32736230-DIC-1.jpg",
"id": 32736232
}
],
"id": 32736230
}
]
Imaging Data Gallery
https://transgeneome.mpi-cbg.de/transgeneomics/api/gallery.jpg?id=[ID]
e.g.:
https://transgeneome.mpi-cbg.de/transgeneomics/api/gallery.jpg?id=32736230
ImagingDataChannel
Having the imaging data channel id from the json, would allow a call like
https://transgeneome.mpi-cbg.de/transgeneomics/api/imagingDataChannel.jpg?id=46472095
which would result in a file like
Linking back to The Transgeneome DB
This will be relevant once the objects are on the site. Ask Sybil or Todd to implement the same mechanism is in place for Wormviz to link back to TransgeneOme
That URL will lead all images/annotations for that particular gene. Use the WBGeneId
https://transgeneome.mpi-cbg.de/transgeneomics/public/geneLocalization.html?geneDbId=WBGene00001689
Generating a .ace file
Tazendra directory /home/azurebrd/work/parsings/daniela/20160324_transgeneome/
The parsing of the JSON to just get data is here : /home/azurebrd/work/parsings/daniela/20160324_transgeneome/all_data
the json output is here: https://transgeneome.mpi-cbg.de/transgeneomics/api/imagingData.json
Parsing output example for gene WBGene00023497 /home/azurebrd/work/parsings/daniela/20160324_transgeneome/WBGene00023497
Expr_pattern : "someID" WBGene : "WBGene00023497" // 31872735 Strain "OP184" Transgene "wgIs184" Remark "Annotated by TransgeneOmics Facility" Anatomy_term "WBbt:0003681" "WBls:0000024" Anatomy_term "WBbt:0005135" "WBls:0000024" Anatomy_term "WBbt:0005741" "WBls:0000024" Anatomy_term "WBbt:0005742" "WBls:0000024" Anatomy_term "WBbt:0006751" "WBls:0000024" Anatomy_term "WBbt:0006761" "WBls:0000024" Anatomy_term "WBbt:0006919" "WBls:0000024" GO_term "GO:0005634"