Difference between revisions of "Pictures"
| (824 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
[[Pictures]]<br> | [[Pictures]]<br> | ||
---- | ---- | ||
| + | ==Picture Data Model== | ||
| + | //////////////////////////////////////////////////////////////////////////////////// | ||
| + | ?Picture Description ?Text | ||
| + | Name UNIQUE Text | ||
| + | Crop Crop_picture ?Picture XREF Cropped_from | ||
| + | Cropped_from ?Picture XREF Crop_picture | ||
| + | Pick_me_to_call Text Text | ||
| + | Remark ?Text #Evidence | ||
| + | Depict Expr_pattern ?Expr_pattern XREF Picture | ||
| + | Anatomy ?Anatomy_term XREF Picture | ||
| + | Cellular_component ?GO_term XREF Picture | ||
| + | Acknowledgment Template UNIQUE Text | ||
| + | Publication_year UNIQUE Text | ||
| + | Article_URL UNIQUE ?Database UNIQUE ?Database_field UNIQUE ?Accession_number | ||
| + | Journal_URL UNIQUE ?Database | ||
| + | Publisher_URL UNIQUE ?Database | ||
| + | Person_name UNIQUE Text | ||
| + | Reference ?Paper XREF Picture | ||
| + | Contact ?Person | ||
| + | |||
| + | |||
| + | /////////////////////////////////////////////////////////////////////////////////// | ||
| + | |||
==Picture Curation== | ==Picture Curation== | ||
The immediate goal of '''picture curation''' is to be able to obtain images of gene expression data from the literature and individual laboratories and display them in the WormBase gene expression page. | The immediate goal of '''picture curation''' is to be able to obtain images of gene expression data from the literature and individual laboratories and display them in the WormBase gene expression page. | ||
| Line 15: | Line 38: | ||
==Pipeline== | ==Pipeline== | ||
| − | In the early phases of curation, pictures will be taken from open access journals (e.g. PLoS). During the process of | + | In the early phases of curation, pictures will be taken from open access journals (e.g. PLoS, BMC, Biomed Central LTD). During the process of open access image curation, other publishers will be contacted for obtaining copyright permissions. |
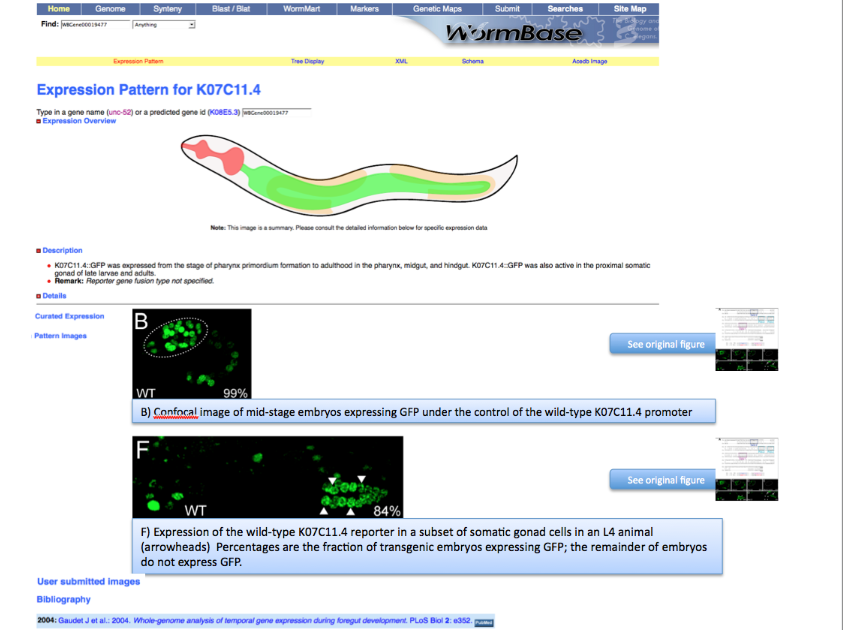
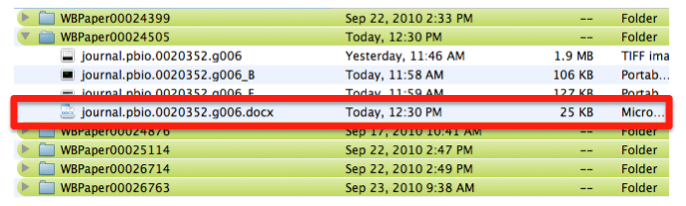
The images should be saved and stored according to the following guidelines. The example shown below refers to a PLoS Biology paper but the rules of handling the pictures are universal and not "paper specific". | The images should be saved and stored according to the following guidelines. The example shown below refers to a PLoS Biology paper but the rules of handling the pictures are universal and not "paper specific". | ||
| Line 34: | Line 57: | ||
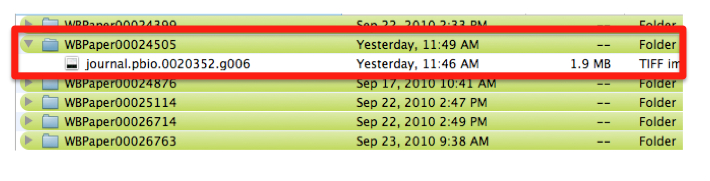
| − | Pictures are saved with their original name in order to minimize editing from the curator. In this case the file is called “journal.pbio.0020352.g006”. | + | Pictures are saved with their original name in order to minimize editing from the curator. In this case the file is called “journal.pbio.0020352.g006”. The files are directly converted into JPEG. TIFF is not indicated as web display format. Avoid using special characters like ' * / in the file name. |
The file is saved in a directory named after the WB paper ID. E.g.: WBPaper00024505, meaning that picture “journal.pbio.0020352.g006” has been downloaded from WBPaper00024505. | The file is saved in a directory named after the WB paper ID. E.g.: WBPaper00024505, meaning that picture “journal.pbio.0020352.g006” has been downloaded from WBPaper00024505. | ||
| Line 75: | Line 98: | ||
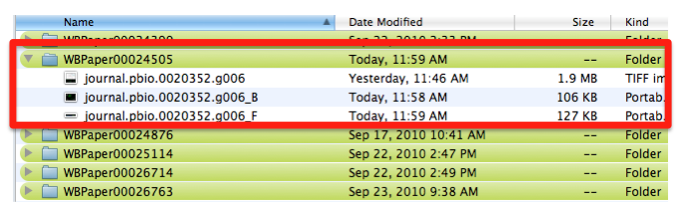
| − | At the same time, the text file associated with the entire figure WBPicture000000001, is saved with the same name as the figure -journal.pbio.0020352.g006- with a . | + | At the same time, the text file associated with the entire figure WBPicture000000001, is saved with the same name as the figure -journal.pbio.0020352.g006- with a .txt extension. In this way we can make sure which figure legend goes with which picture. |
| − | |||
[[File:PictureE1.png]] | [[File:PictureE1.png]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
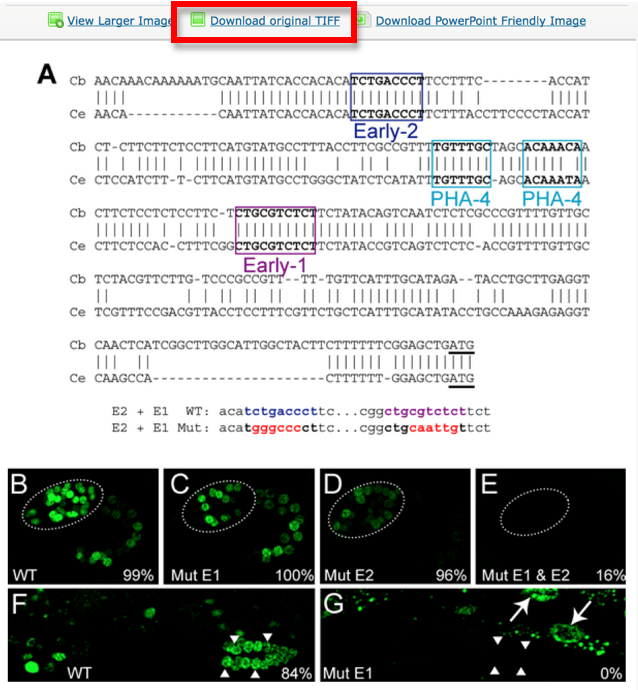
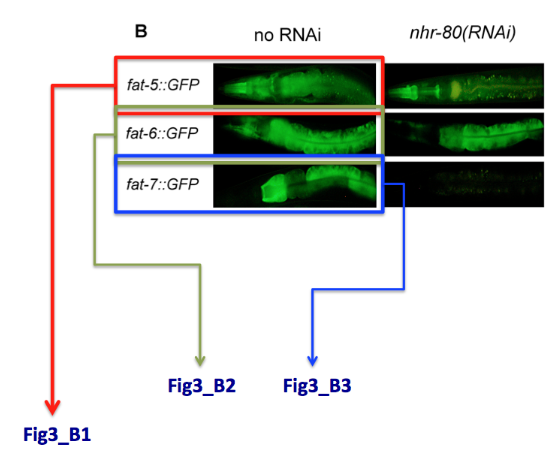
'''Special case:''' what do I do when one single panel refers to multiple genes. E.g. In the example below, panel B displays the expression of 3 different genes. We will simply name the pictures Fig3_B1, Fig3_B2, Fig3_B3. | '''Special case:''' what do I do when one single panel refers to multiple genes. E.g. In the example below, panel B displays the expression of 3 different genes. We will simply name the pictures Fig3_B1, Fig3_B2, Fig3_B3. | ||
| Line 120: | Line 132: | ||
[[File:PictureL.png]] | [[File:PictureL.png]] | ||
| − | ''' | + | '''To the web team: in this case is the [[Picture Data Model]] proposed sufficient to determine that this picture should be displayed as PARENTAL or DAUGHTER?''' Yes |
| + | |||
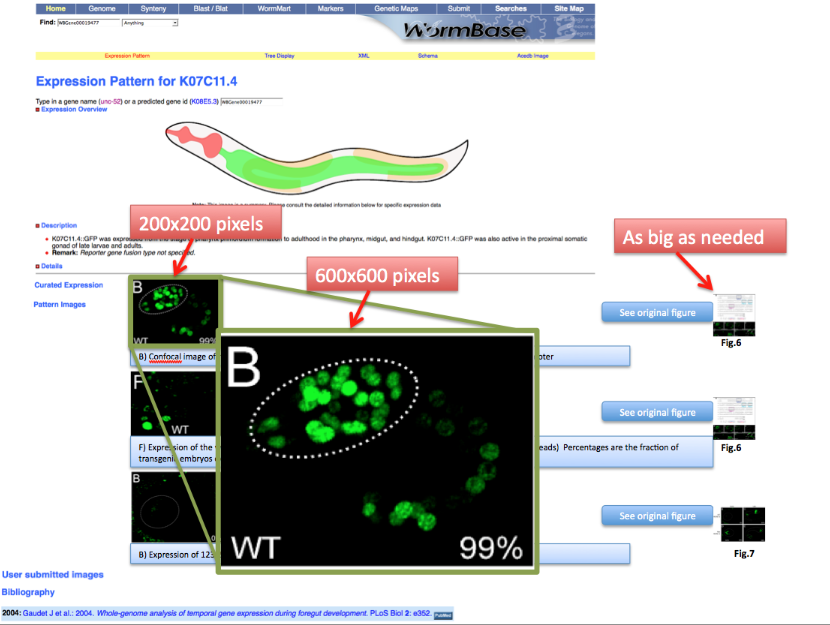
| + | ===Picture size and format=== | ||
| + | |||
| + | All the pictures should be in JPEG format, if possible. | ||
| + | |||
| + | The picture size for thumbnails shown in the main gene expression page should be 200x200 pixels. | ||
| + | |||
| + | Picture size for the full view 600x600 pixels. | ||
| + | |||
| + | Picture size for the original file will be as big as needed. | ||
| + | |||
| + | NB: a note on 200x200 and 600x600 pixel size. This will not distort the pictures but just put a constraint on the maximum size of the thumbnail or the full image. | ||
| + | |||
| + | [[File:PictureM1.png]] | ||
| + | |||
| + | ===Summary Pipeline for picture handling=== | ||
| + | * Pictures are saved and organized in folders in Lario (Daniela's computer) as described above. The folder name will be the WBPaperID or the WBPersonID | ||
| + | |||
| + | For picture download there are 2 main pipelines: | ||
| + | * 1- fetch picture with the script fetchpictures.pl | ||
| + | * 2- fetch pictures via Arun's script | ||
| + | |||
| + | After downloading the pictures move them in the OICR folder (/Users/danielaraciti/Desktop/Canopus/Pictures/OICR). | ||
| + | |||
| + | * Run Juancarlos' script to generate the picture_source file: ./mergeToOICR.pl | ||
| + | |||
| + | * Go to /Users/danielaraciti/Desktop/Canopus/Pictures and scp the file that was generated - picture_source- to tazendra scp picture_source acedb@tazendra.caltech.edu:/home/acedb/draciti/picture_source/ | ||
| + | |||
| + | * rsync the file to Canopus rsync --progress --delete-after -a /Users/danielaraciti/Desktop/Canopus/Pictures/OICR/ daniela@canopus.caltech.edu:OICR/ | ||
| + | |||
| + | * Go to canopus and run the script /home/daniela/OICR/makethumb.sh. the script will now generate 200 and 600 thumbnails. | ||
| + | |||
| + | |||
| + | To update the term info display into picture OA on tazendra: | ||
| + | |||
| + | Go to | ||
| + | /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl | ||
| + | |||
| + | this script updates directly the term info display into the OA> You have now links to the new pictures. | ||
| + | |||
| + | |||
| + | Canopus Accessibility: | ||
| + | |||
| + | '''this is the location from where OICR takes images and movies''' | ||
| + | '''ftp://caltech.wormbase.org/pub/OICR/''' | ||
| + | |||
| + | |||
| + | on canoups every night, /home/daniela/OICR/* is copied over to /usr/local/wormbase/OICR/ and /srv/ftp/pub/OICR/ (where files are | ||
| + | accessible via ftp) | ||
| + | |||
| + | |||
| + | Example for movies | ||
| + | canopus.caltech.edu:/usr/local/wormbase/OICR/Movies | ||
| + | ftp://canopus.caltech.edu/pub/OICR/Movies | ||
| + | |||
| + | |||
| + | OICR file location: | ||
| + | |||
| + | /usr/local/wormbase/website-shared-file/html/img-static/movies | ||
| + | |||
| + | ====Naming folders after WBPersonID==== | ||
| + | |||
| + | When you have multiple contacts remember to call the folder where you store pictures with the WBPersonID of the first person inserted in the OA | ||
| + | |||
| + | ====Large Scale imports==== | ||
| + | |||
| + | When you import a large scale study remember to put only the contact and person and to put the publication in the remarks as a brief citation. This is to prevent conflicts in the picture fetching process from OICR. The pictures will be called after Paper OR Person. | ||
| + | |||
| + | ====Pictures added by other curators==== | ||
| + | a cronjob is checking daily if there are new objects added by other curators | ||
| + | the cronjob is set on tazendra in the acedb account | ||
| + | # 0 1 * * * /home/acedb/draciti/cronjobs/daily_pictures_curator.pl | ||
| + | so every day at 1am | ||
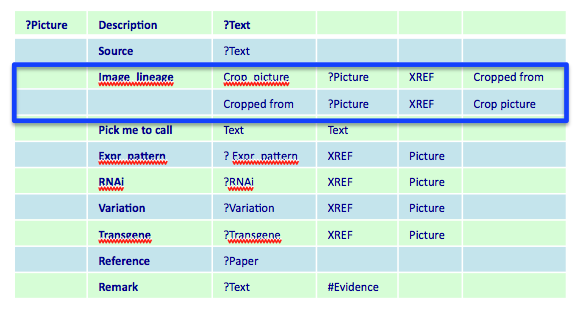
==Picture Data Model Proposal== | ==Picture Data Model Proposal== | ||
//////////////////////////////////////////////////////////////////////////////////// | //////////////////////////////////////////////////////////////////////////////////// | ||
?Picture Description ?Text | ?Picture Description ?Text | ||
| − | + | Name UNIQUE Text | |
| − | + | Crop Crop_picture ?Picture XREF Cropped_from | |
| − | + | Cropped_from ?Picture XREF Crop_picture | |
Pick_me_to_call Text Text | Pick_me_to_call Text Text | ||
| − | Expr_pattern ?Expr_pattern XREF Picture | + | Remark ?Text #Evidence |
| + | Depict Expr_pattern ?Expr_pattern XREF Picture | ||
| + | Anatomy ?Anatomy_term XREF Picture | ||
| + | Cellular_component ?GO_term XREF Picture | ||
| + | Acknowledgment Template UNIQUE Text | ||
| + | Publication_year UNIQUE Text | ||
| + | Article_URL UNIQUE ?Database UNIQUE ?Database_field UNIQUE ?Accession_number | ||
| + | Journal_URL UNIQUE ?Database | ||
| + | Publisher_URL UNIQUE ?Database | ||
| + | Person_name UNIQUE Text | ||
Reference ?Paper XREF Picture | Reference ?Paper XREF Picture | ||
| − | + | Contact ?Person | |
| − | + | ||
| − | + | ||
/////////////////////////////////////////////////////////////////////////////////// | /////////////////////////////////////////////////////////////////////////////////// | ||
==Picture Data Model step by step explanation== | ==Picture Data Model step by step explanation== | ||
| − | + | '''Picture''' | |
Name of the picture object. E.g. WBPicture0000000001 | Name of the picture object. E.g. WBPicture0000000001 | ||
| − | + | '''Description''' | |
Figure legend | Figure legend | ||
| − | + | '''Name''' | |
| − | For actual picture names. This is the name of the path leading to the picture file. The source includes the name of the directory where the picture comes from AND the name of the picture file. e.g. WBPaper00024505_journal.pbio.0020352.g006 | + | For actual picture names. (This is the name of the path leading to the picture file. The source includes the name of the directory where the picture comes from AND the name of the picture file. e.g. WBPaper00024505_journal.pbio.0020352.g006. Deprecated). New decision made with web team. The "name" will only be the name of the file e.g.: journal.pbio.0020352.g006. The web team will construct the path as we discussed via e-mail on Dec 8th: |
| − | + | "So to back this up we will provide in the .ace file | |
| + | |||
| + | Reference "WBPaper12345678" | ||
| + | |||
| + | Contact "WBPerson1234" | ||
| + | |||
| + | Name "pic.abcdefg.jpg" | ||
| + | |||
| + | and the rule to construct the path is: | ||
| + | |||
| + | knowing that if there is a reference the path will be Reference/Name (e.g. WBPaper12345678/pic.abcdefg.jpg) | ||
| + | and if there is no Reference it will be Contact/Name (WBPerson1234/pic.abcdefg.jpg)" Daniela | ||
| + | |||
| + | '''Crop''' | ||
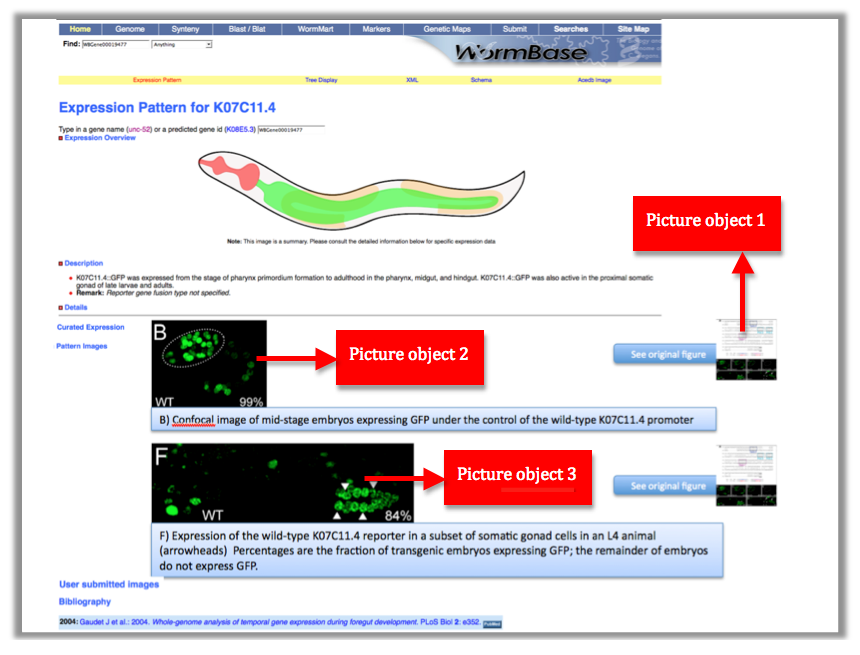
This is the picture object lineage. Large figures will be cropped into sections when they represent different data. We want to maintain the picture lineage -> by clicking on the | This is the picture object lineage. Large figures will be cropped into sections when they represent different data. We want to maintain the picture lineage -> by clicking on the | ||
"see original figure button" we want to access the entire image. | "see original figure button" we want to access the entire image. | ||
| − | + | '''Pick_me_to_call''' | |
| − | + | Untouched tag from the existing model. | |
| + | |||
| + | '''Remark''' | ||
| + | For curator notes | ||
| − | + | Depict | |
| − | |||
| − | + | '''Expr_pattern''' | |
| − | For the | + | For linking to Expr-pattern data. This will be the Expr_pattern object that is associated with the picture. |
| − | + | '''Anatomy''' | |
| − | + | It will link the picture object directly to an Anatomy Object | |
| − | |||
| − | + | '''Cellular_component''' | |
This links to the GO term e.g. if a picture depicts sub-cellular localization | This links to the GO term e.g. if a picture depicts sub-cellular localization | ||
| − | ===Anatomy object=== | + | |
| − | It will link the picture object directly to an Anatomy Object | + | '''Reference''' |
| + | For the source of the picture E.g.WBPaper12345678. | ||
| + | |||
| + | '''Contact''' | ||
| + | Whenever the picture does not come from a publication but from a person/lab this is the person who should be contacted. Normally the PI of the lab where the picture has been generated. | ||
| + | |||
| + | Note: the following tags were removed from the model: | ||
| + | |||
| + | RNAi | ||
| + | |||
| + | Variation | ||
| + | |||
| + | Transgene | ||
| + | |||
| + | because there were no data associated to those tags. The search to check association was done last time on November 4th on WS219 | ||
| + | |||
| + | ==='''Acknowledgment'''=== | ||
| + | |||
| + | e.g.: | ||
| + | WormBase thanks the journal Genetics <http://www.genetics.org/> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from Chen et al, Genetics 166:151-60, sel-7, a positive regulator of lin-12 activity, encodes a novel nuclear protein in Caenorhabditis elegans <http://www.genetics.org/cgi/content/full/166/1/151>. Copyright (2004) with permission from the Genetics Society of America <http://www.genetics-gsa.org/>. | ||
| + | |||
| + | In the sentence there are 4 variables: | ||
| + | |||
| + | "WormBase thanks the journal <Journal_URL> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Article_URL>. Copyright (<Publication_year>) with permission from <Publisher_URL>." | ||
| + | |||
| + | |||
| + | |||
| + | * Acknowledgment UNIQUE Template Text | ||
| + | * Publication_year UNIQUE Text | ||
| + | * Journal_URL UNIQUE ?Database | ||
| + | * Article_URL UNIQUE ?Database UNIQUE ?Database_field UNIQUE ?Accession_number | ||
| + | * Publisher_URL UNIQUE ?Database | ||
| + | * Person_name Text | ||
| + | |||
| + | |||
| + | where | ||
| + | |||
| + | '''Template''' | ||
| + | Is the template sentence e.g. "WormBase thanks the journal <Journal_URL> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Article_URL>. Copyright (<Publication_year>) with permission from <Publisher_URL>." | ||
| + | |||
| + | The template sentence will change accordingly to what publishers need but the tags populating it will always be the ones listed below | ||
| + | |||
| + | '''Publication_year''' | ||
| + | self explanatory | ||
| + | |||
| + | '''Journal_URL''' | ||
| + | this will contain the URL pointing to the journal home page. | ||
| + | |||
| + | '''Article_URL''' | ||
| + | this will contain the URL pointing to the paper citation. | ||
| + | |||
| + | '''Publisher_URL''' | ||
| + | this will contain the URL pointing to the publisher's homepage. | ||
| + | |||
| + | '''Person_name''' | ||
| + | if the picture is given by a person/lab | ||
| + | |||
| + | ====Update October 2023==== | ||
| + | *** To allow consistency in links and to avoid broken links due to publisher's changes we did update the Article_URL so that it will point to the DOI. See this ticket for more information -https://github.com/WormBase/website/issues/9349. | ||
| + | |||
| + | ==Example== | ||
| + | |||
| + | Picture : "WBPicture0000000001" | ||
| + | |||
| + | Description "Figure Legend: A. ..... B. ..... C. .... D ....."" | ||
| + | |||
| + | Name "journal.pbio.0020352.g006_B" | ||
| + | |||
| + | Cropped_from "journal.pbio.0020352.g006" | ||
| + | |||
| + | Remark "Some remark" | ||
| + | |||
| + | Expr_pattern "Expr1234" | ||
| + | |||
| + | Anatomy "WBbt:0004017" | ||
| + | |||
| + | Cellular_component "GO:0005634" | ||
| + | |||
| + | Template "WormBase thanks the journal <Journal_URL> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Journal_URL>, <Article_URL>. Copyright <Publication_year> with permission from <Publisher_URL>." | ||
| + | |||
| + | Publication_year "2004" | ||
| + | |||
| + | Article_URL WBPaper00024505_URL id 0020352 | ||
| + | |||
| + | Journal_URL "PLoSBiology" | ||
| + | |||
| + | Publisher_URL "PLoS" | ||
| + | |||
| + | Reference "WBPaper00024505" | ||
| + | |||
| + | Contact "WBPerson123" | ||
| + | |||
| + | Database : WBPaper00024505_URL | ||
| + | |||
| + | Name "Ding M et al. (2008) PLoS One \"The cell signaling adaptor protein EPS-8 is essential for C. elegans epidermal ....\"" | ||
| + | |||
| + | URL_constructor "http:\/\/www.plosbiology.org\/article\/info:doi%2F10.1371%2Fjournal.pbio.%S" | ||
| + | |||
| + | Database : PLoSBiology | ||
| + | |||
| + | Name "PLoS Biology" | ||
| + | |||
| + | URL_constructor "http:\/\/www.plosbiology.org\/" | ||
| + | |||
| + | Database : PLoS | ||
| + | |||
| + | Name "PLoS" | ||
| + | |||
| + | URL_constructor "http:\/\/www.plos.org\/" | ||
| + | |||
| + | == OA interface== | ||
| + | ===Tab1=== | ||
| + | *''Pgdbid'' - postgres database ID, generates automatically upon entry. | ||
| + | *''WBPicture'' - Generates automatically upon entry. When duplicating a picture object be sure to assign a new Picture ID number which has the same number as the postgres ID (pgid). Be extra careful when you change name to an existing object. See section ''Renaming objects''. | ||
| + | *''Reference'' - WBPaperID paper ontology - | ||
| + | *''Contact'' - Multiontology on people. N.B. when you have more than one Contact be aware on how to name the folder that stores pictures. As a general rule the folder should always be called after the first person inserted. | ||
| + | *''Description'' - Figure legend (bigtext) | ||
| + | *''Source'' - Multiontology from file picture_source on Tazendra (acedb@tazendra.caltech.edu:/home/acedb/draciti/) | ||
| + | *''Cropped_From'' - Multiontology on source then WBPicture name/ID. Show on Term Info name/ID, source, reference. | ||
| + | *''Expr_pattern'' - source file in Tazendra: /home/acedb/draciti/ExprWS221.ace. In term info we'd like to see Gene, Pattern, Reference, Reporter_gene, Life_stage Anatomy_term, GO_term. Autocomplete just on Expr_pattern ID. | ||
| + | *''Topic'' - multiontology on process term | ||
| + | *''Remark'' - bigtext | ||
| + | *''Cellular component'' - multiontology of GO_Term like gop_goid. source file in Tazendra: /home/acedb/draciti/ExprWS221.ace. | ||
| + | *''Anatomy term'' - multiontology. Should work like app_anatomy. source file in Tazendra: /home/acedb/draciti/ExprWS221.ace. File that has Anatomy_term <-> anatomy name association is https://github.com/raymond91125/Wao/raw/master/WBbt.obo. Previously was: http://brebiou.cshl.edu/viewcvs/*checkout*/Wao/WBbt.obo but became obsolete 04-11-2011 DR (and even before was http://obo.cvs.sourceforge.net/viewvc/obo/obo/ontology/anatomy/gross_anatomy/animal_gross_anatomy/worm/worm_anatomy/WBbt.obo but became obsolete 02-11-2011 DR) | ||
| + | *''URL_Accession'' - text | ||
| + | *''Person'' - Multiontology on people | ||
| + | *''Person_text'' - free text. The person text was created for cases when we want to acknowledge people that are not WBPersons. Note on the .ace file for Person and Person_name: join all <pic_person> objects's starndard names with commas, then comma, then <pic_persontext> text. If there's no <pic_persontext>: join all person objects's standard names with commas, except for the last one, which is joined by "<comma> and ". Daniela remember for curation when you enter a text in the person text you should do the following: If it is a single entity in the text write e.g. "and Paul Sternberg's lab" if there are multiple entities e.g. summer student 1 (non WBPerson), summer student 2 (non WBPerson), and summer student 3 (non WBPerson) write "John Smith, George Brown, and Mike Lee". It is constructed in this way otherwise the syntax would have been too complicated | ||
| + | *''Life_stage'' - like in the phenotype OA | ||
| + | *''Curator'' - Multiontology on people | ||
| + | *''No dump'' - Toggle | ||
| + | *''Chris Flag'' - Toggle | ||
| + | '''*''Permission'' - Dropdown with the following values: blank|Daniela|e-mail sent|granted|rejected''' | ||
| + | |||
| + | *''Acknowledgment'' - the acknowledgment section is hard coded in the dumper script. | ||
| + | '''modify the dumper for acknowledgment for PNAS -not urgent, need to be ready when starting annotating PNAS papers. We need two different acknowledgments: till end of 2008 use this acknowledgment: Reprinted with permission from <Journal_URL> <Article_URL>. Copyright (<Publication_year>), <Publisher_URL>. From 2009 on use this: Reprinted with permission from <Journal_URL> <Article_URL>, <Publisher_URL>.''' | ||
| + | |||
| + | If there is a Reference the Acknowledgment is constructed this way: The information on Journal_name will be taken from Paper tables and If the Journal_name is empty write BLANK. The same is true for Publication_year, will be taken from Paper tables and If the Journal_name is empty write BLANK. The mapping file for the other fields of the acknowledgments is called Mapings.txt and is on Tazendra: /home/acedb/draciti/Mappings.txt. The file contains The template text, Article_URL, Journal_URL, and Publisher_URL. | ||
| + | |||
| + | When there is no Reference but there is a Contact the Acknowledgment will also be constructed automatically. Wormbase thanks <Person_name> for providing the pictures. The Person_name is constructed with the "Person" and "Person_text" tags (hardcoded in the dumper script). See details in the dumper section. | ||
| + | |||
| + | If a paper has a PMID and is missing Journal or Year, let Kimberly know. If it doesn't have a PMID and is missing that information, fill it in using the paper editor. | ||
| + | |||
| + | ===tab2=== | ||
| + | The postgres tables are | ||
| + | *''Phenotype'' multiontology on phenotypes pic_phenotype | ||
| + | *''allele''ontology on variations pic_variation | ||
| + | *''Gene'' multiontology on genes pic_wbgene | ||
| + | |||
| + | == TODO Daniela when generating Expr_pattern OA == | ||
| + | Other OA configs going to use WBPicture object ID: Expr_pattern OA (when it will exist) | ||
| + | When coding complete: | ||
| + | J set milestone to "code complete" | ||
| + | D check that it works, set milestone to "verified" | ||
| + | J make it live on tazendra, set milestone to "live" | ||
| + | D check it works, and resolve the issue. | ||
| + | |||
| + | == mapping of OA fields to postgres tables == | ||
| + | |||
| + | * WBPicture -> pic_name | ||
| + | * Reference -> pic_paper | ||
| + | * Contact -> pic_contact | ||
| + | * Description -> pic_description | ||
| + | * Source -> pic_source | ||
| + | * Cropped_from -> pic_croppedfrom | ||
| + | * Expression Pattern -> pic_exprpattern | ||
| + | * Topic -> pic_process | ||
| + | * Remark -> pic_remark | ||
| + | * Cellular_component -> pic_goid | ||
| + | * Anatomy_term -> pic_anat_term | ||
| + | * URL Accession -> pic_urlaccession | ||
| + | * Person -> pic_person | ||
| + | * Person Text -> pic_persontext | ||
| + | * Life Stage -> pic_lifestage | ||
| + | * Species -> pic_species | ||
| + | * Curator -> pic_curator | ||
| + | * NO DUMP -> pic_nodump | ||
| + | * Chris Flag -> pic_chris | ||
| + | * Phenotype -> pic_phenotype | ||
| + | * Allele -> pic_variation | ||
| + | * Gene -> pic_wbgene | ||
| + | |||
| + | ==Test dumper script== | ||
| + | |||
| + | On the Sandbox: | ||
| + | |||
| + | go to mangolassi | ||
| + | |||
| + | ''ssh acedb@mangolassi.caltech.edu'' | ||
| + | |||
| + | ''cd /home/acedb/draciti/oa_picture_ace_dumper'' | ||
| + | |||
| + | ''./dump_picture_ace.pl'' | ||
| + | |||
| + | the dumper generates 2 files: picture.ace and pictures.err | ||
| + | |||
| + | |||
| + | On Tazendra: | ||
| + | |||
| + | go to Tazendra | ||
| + | |||
| + | ''ssh acedb@tazendra.caltech.edu'' | ||
| + | |||
| + | ''cd /home/acedb/draciti/oa_picture_ace_dumper'' | ||
| + | |||
| + | ''./dump_picture_ace.pl'' | ||
| + | |||
| + | == Chronograms == | ||
| + | |||
| + | The chronogram could not be put in the Expr_pattern field. The Chronogram name will be put in directly frorm the dumper script. Juancarlos could not fit in Chronograms in the ontology as that applied only to Expr_patterns. In the .ace file we have anyway the tag "Expr_pattern "Chronogram1"" so the association in the picture page should display fine. | ||
| + | |||
| + | == .ace template for dumping== | ||
| + | |||
| + | ''Picture'' : <pic_name> | ||
| + | |||
| + | ''Description'' "<pic_description>" | ||
| + | |||
| + | ''Name'' "<pic_source>" | ||
| + | |||
| + | ''Cropped_from'' "<pic_croppedfrom>" | ||
| + | |||
| + | ''Remark'' "<pic_remark>" | ||
| + | |||
| + | ''Expr_pattern'' "<pic_exprpattern>" | ||
| + | |||
| + | ''Anatomy'' "<pic_anat_term>" | ||
| + | |||
| + | ''Cellular_component'' "<pic_goid>" | ||
| + | |||
| + | ''Template'' "<Template Text>" from Mappings.txt file on tazendra. When there is no Reference but there is a Contact it will generate automatically the template sentence Wormbase thanks <Person_name> for providing the pictures (hard-coded in the dumper). | ||
| + | |||
| + | ''Publication_year'' -- take this from Paper tables. Juancarlos I don't know the specifics of the Paper tables | ||
| + | |||
| + | ''Article_URL'' "<pic_paper>_URL id <pic_urlaccession>" | ||
| + | |||
| + | ''Journal_URL'' "<Full Journal Name>" from the Mappings.txt file on tazendra | ||
| + | |||
| + | ''Publisher_URL'' "<Publisher_name>" from Mappings.txt file on tazendra | ||
| + | |||
| + | ''Reference'' "<pic_paper>" | ||
| + | |||
| + | ''Contact'' "<pic_contact>" | ||
| + | |||
| + | |||
| + | ''Database'' : "<pic_paper>_URL" | ||
| + | |||
| + | ''Name'' - take this from Brief citation from Paper model NB There are "" that have to be escaped with backslash \ otherwise the .ace file is not reading in fine. Brief_citation name coming from new module at /home/postgres/work/citace_upload/papers/get_brief_citation.pm -- J | ||
| + | |||
| + | ''URL_constructor'' "<Article_URL>" from Mappings.txt file on tazendra | ||
| + | |||
| + | |||
| + | |||
| + | ''Database'' : "<Full Journal Name>" from Mappings.txt file on tazendra | ||
| + | |||
| + | ''Name'' "<Full Journal Name>" from Mappings.txt file on tazendra | ||
| + | |||
| + | ''URL_constructor'' "<Journal_URL>" from Mappings.txt file on tazendra | ||
| + | |||
| + | |||
| + | |||
| + | ''Database'' : "<Publisher_name>" in Mappings.txt file on tazendra | ||
| + | |||
| + | ''Name'' "<Publisher_name>" in Mappings.txt file on tazendra | ||
| + | |||
| + | ''URL_constructor'' "<Publisher_URL>" from Mappings.txt file on tazendra | ||
| + | |||
| + | |||
| + | |||
| + | .ace dumper at mangolassi at /home/acedb/draciti/oa_picture_ace_dumper/ | ||
| + | (actually at /home/postgres/work/citace_upload/picture/ and symlinked here) | ||
| + | |||
| + | called dump_picture_ace.pl | ||
| + | |||
| + | generates pictures.ace and pictures.err (errorfile, always look at this even if it's usually empty) -- J | ||
| + | |||
| + | ==How the dumper works== | ||
| + | |||
| + | Any error will be written in pictures.err in /home/acedb/draciti/oa_picture_ace_dumper/ Daniela always check it. | ||
| + | Any output is going to be in pictures.ace in /home/acedb/draciti/oa_picture_ace_dumper/ | ||
| + | |||
| + | An error will be written if there are 2 ore more pgid for the same Paper+source. | ||
| + | |||
| + | |||
| + | the first thing the dumper does is to read into the Picture_source file /home/acedb/draciti/picture_source/picture_source and puts that in a hash %urlacc | ||
| + | If the file is ever not there, the script won't work. | ||
| + | |||
| + | my %urlacc; | ||
| + | &readUrlacc(); | ||
| + | sub readUrlacc { | ||
| + | my $infile = '/home/acedb/draciti/picture_source/picture_source'; | ||
| + | open (IN, "<$infile") or die "Cannot open $infile : $!"; | ||
| + | while (my $line = <IN>) { | ||
| + | my ($paper, $filename, $urlaccession) = split/\t/, $line; | ||
| + | if ($urlaccession) { $urlacc{$paper} = $urlaccession; } | ||
| + | } # while (my $line = <IN>) | ||
| + | close (IN) or die "Cannot close $infile : $!"; | ||
| + | } # sub readUrlacc | ||
| + | |||
| + | if it finds an association with the WBPaperID and the URL_accession it prints the URL accession | ||
| + | then it reads the Mappings.txt file on mangolassi (later on Tazendra) /home/acedb/draciti/oa_picture_ace_dumper/ | ||
| + | This file contains the mappings publisher | ||
| + | the script will skip the 1st line cause is the header. | ||
| + | For each of every other line: | ||
| + | |||
| + | splits into tabs to get each field. | ||
| + | |||
| + | 1 $pubname (1st column) | ||
| + | |||
| + | 2 $puburl (2nd column) | ||
| + | |||
| + | 3 $journame (3rd column) | ||
| + | |||
| + | 4 $jourfull (4th column) | ||
| + | |||
| + | 5 $joururl (5th column) | ||
| + | |||
| + | 6 $arturl (6th column) | ||
| + | |||
| + | 7 $template (7th column) | ||
| + | |||
| + | If there is no journal name it skips the line. Each of the values in the Mappings.txt file is associated to the Journal name (3rd column). Additionally, for the Database field, we need a Full Journal Name without spaces associated to the Journal Name otherwise will not read into acedb. | ||
| + | |||
| + | For those 7 values if any of them is missing it will give an error line. | ||
| + | |||
| + | It creates | ||
| + | |||
| + | $entry = "Database : \"$stripped_pubname\"\n"; | ||
| + | $entry .= "Name\t\"$pubname\"\n"; | ||
| + | $entry .= "URL_constructor\t\"$puburl\"\n"; | ||
| + | $entry .= "\n"; | ||
| + | |||
| + | Juancarlos, I would like to escape any "/" with a "\" for the following columns of the mappings.txt file. 2nd column: $puburl, 5th column: $joururl, 6th column: $arturl. I have already modified the Mappings.txt file on Mangolassi. J Done | ||
| + | |||
| + | which will be displayed only once at the beginning of the .ace file. | ||
| + | |||
| + | e.g.: Database : "PLoS" | ||
| + | |||
| + | Name "PLoS" | ||
| + | |||
| + | URL_constructor "http:\/\/www.plos.org\/" | ||
| + | |||
| + | |||
| + | $entry = "Database : \"$stripped_jourfull\"\n"; | ||
| + | $entry .= "Name\t\"$jourfull\"\n"; | ||
| + | $entry .= "URL\t\"$joururl\"\n"; | ||
| + | $entry .= "URL_constructor\t\"$arturl\"\n"; | ||
| + | $entry .= "\n"; | ||
| + | |||
| + | also displayed only once at the beginning of the .ace file. | ||
| + | |||
| + | Database : "PLoSBiology" | ||
| + | |||
| + | Name "PLoS Biology" | ||
| + | |||
| + | URL "http:\/\/www.plosbiology.org\/" | ||
| + | |||
| + | URL_constructor "http://www.plosbiology.org/article/info:doi%2F10.1371%2Fjournal.pbio.%S" //added 02-10-2011 Daniela&Juancarlos | ||
| + | |||
| + | This is it for reading the Mappings.txt file | ||
| + | |||
| + | |||
| + | |||
| + | list of postgres tables: | ||
| + | anat_term goid nodump persontext urlaccession chris description lifestage paper remark croppedfrom exprpattern name person source contact | ||
| + | |||
| + | each of the OA fields in the list maps to a pic_ table (see mapping of OA fields to postgres tables chapter in wiki for mappings) | ||
| + | |||
| + | At this point we read the data from postgres as long as there are data in the table for each entry. | ||
| + | |||
| + | After reading into the data it replaces all the new line (line breaks) with a space (e.g. in the remark field if I enter text in separate lines it will display it in the .ace file on the same line) | ||
| + | |||
| + | Then it is creating a mapping from PersonIDs to standard names. | ||
| + | |||
| + | For each paper we are also getting a mapping of the PaperID to | ||
| + | |||
| + | journal | ||
| + | |||
| + | year | ||
| + | |||
| + | title | ||
| + | |||
| + | the first author (if there is also a second author it will add et al.) | ||
| + | |||
| + | ===starting to dump=== | ||
| + | |||
| + | The dumper looks at all entries that have a PictureID and for each of them it does the following: | ||
| + | |||
| + | get its pgid. If that pgid has a NO DUMP value it will skip it. | ||
| + | |||
| + | then it will create 2 objects (at the same time): | ||
| + | |||
| + | - Picture object | ||
| + | |||
| + | - Database object | ||
| + | |||
| + | Then the dumper will: | ||
| + | |||
| + | $entry .= "Picture : \"$data{name}{$pgid}\"\n"; | ||
| + | if ($data{description}{$pgid}) { $entry .= "Description\t\"$data{description}{$pgid}\"\n"; } | ||
| + | if ($data{source}{$pgid}) { $entry .= "Name\t\"$data{source}{$pgid}\"\n"; } | ||
| + | if ($data{croppedfrom}{$pgid}) { $entry .= "Cropped_from\t\"$data{croppedfrom}{$pgid}\"\n"; } | ||
| + | if ($data{remark}{$pgid}) { $entry .= "Remark\t\"$data{remark}{$pgid}\"\n"; } | ||
| + | |||
| + | meaning if there is a picture object will create a picture object header | ||
| + | |||
| + | e.g. Picture : "WBPicture0000000004" | ||
| + | |||
| + | Juancarlos could you please add the following rule? if pgid and WBPicture number don't match numerically should give an error. Thanks D | ||
| + | |||
| + | when dumping an object you'll get an error if the name has spaces in the front or at the end (very important, because other objects linked to this will also have that space, so you should remove any connections to that object ID, fix the spaces, then remake the connections to the fixed object ID), and/or if the number of the object id isn't the same as the pgid -- J | ||
| + | |||
| + | Juancarlos could you please add the following rule? What is in the "Source" field should be unique. There should never be 2 objects with the same source. D Done, there's already one error of that type, and 2 errors of the pgid not matching object ID type -- J Great thanks! Work fine D | ||
| + | |||
| + | |||
| + | if there is description it will create a Description .ace tag | ||
| + | |||
| + | e.g.: Description "Confocal images of mid-stage embryos expressing GFP under the control of the wild-type K07C11.4 promoter. Percentages are the fraction of transgenic embryos expressing GFP; the remainder of embryos do not express GFP. Dashed lines indicate the outline of the developing pharynx." | ||
| + | |||
| + | Added the following rule: if there is more than one space it will be converted in a single space. Line breaks are deleted. 02.09.2011 | ||
| + | |||
| + | "" symbols are escaped: from J: I'm not escaping it on everything, because the multiontology fields | ||
| + | use "," to separate different values, so it's easier to not mess with | ||
| + | it. Given that they're controlled vocabulary, I've made it filter on | ||
| + | description, source, remark, and urlaccession (not persontext, but we | ||
| + | could add it there, it was just additionally extra code more than the | ||
| + | rest, and I wasn't sure it was necessary). | ||
| + | |||
| + | |||
| + | same for source, cropped_from and remark. | ||
| + | |||
| + | if ($data{exprpattern}{$pgid}) { | ||
| + | my ($data) = $data{exprpattern}{$pgid} =~ m/^\"(.*)\"$/; | ||
| + | my (@data) = split/\",\"/, $data; | ||
| + | foreach my $data (@data) { $entry .= "Expr_pattern\t\"$data\"\n"; } } | ||
| + | if ($data{goid}{$pgid}) { | ||
| + | my ($data) = $data{goid}{$pgid} =~ m/^\"(.*)\"$/; | ||
| + | my (@data) = split/\",\"/, $data; | ||
| + | foreach my $data (@data) { $entry .= "Cellular_component\t\"$data\"\n"; } } | ||
| + | if ($data{anat_term}{$pgid}) { | ||
| + | my ($data) = $data{anat_term}{$pgid} =~ m/^\"(.*)\"$/; | ||
| + | my (@data) = split/\",\"/, $data; | ||
| + | foreach my $data (@data) { $entry .= "Anatomy\t\"$data\"\n"; } } | ||
| + | |||
| + | it does the same as above with the difference that every anatomy entry (or GO entry, or Expr_pattern) will be displayed in a separate line: | ||
| + | e.g: | ||
| + | |||
| + | Anatomy "WBbt:0003681" | ||
| + | |||
| + | Anatomy "WBbt:0005175" | ||
| + | |||
| + | It looks at the contact info, it dumps the Contact e.g. Contact "WBPerson12345" | ||
| + | |||
| + | If more than one contact does the same as Anatomy (1 entry per line). | ||
| + | |||
| + | If there is a contact -> Then it generates a template text: "WormBase thanks <Person_name> for providing the pictures." NB: remember that the dumper will use the Person_name to generate the acknowledgement, not the Contact field. Whenever pictures are submitted from persons other then publications the contact field and the person field should both be filled. (DR 110324) | ||
| + | |||
| + | Now the dumper looks into Person data | ||
| + | |||
| + | if there is a Person and Person Text -> it will join all <pic_person> objects's starndard names with commas, then comma, then <pic_persontext> text. During the curation: If the Person field is filled and you need to add something in the Person Text you should do the following: If it is a single entity in the text write e.g. "and Paul Sternberg's lab" if there are multiple entities e.g. summer student 1 (non WBPerson), summer student 2 (non WBPerson), and summer student 3 (non WBPerson) write "John Smith, George Brown, and Mike Lee". | ||
| + | |||
| + | |||
| + | otherwise if there is person data will | ||
| + | |||
| + | For one person: Juancarlos | ||
| + | for 2 people: Daniela and Juancarlos | ||
| + | For 3 or more people: Daniela, Juancarlos, and Jim | ||
| + | |||
| + | This is the same mapping as before when it converts WBPersonID into standard name | ||
| + | |||
| + | Otherwise if Person_text writes only the person text. Free text. For one person: John Smith. For 2 persons: John Smith and Mark Brown. And so on. | ||
| + | |||
| + | If there is a Reference (pic_paper table in postgres) it dumps the Reference: | ||
| + | |||
| + | e.g.: Reference "WBPaper00024505" | ||
| + | |||
| + | then if there is a year it will generate a Publication_year: | ||
| + | |||
| + | e.g. Publication_year "2004" | ||
| + | |||
| + | If it does not have a publication year it will generate an error (Daniela will fix it according to Kimberly's rules) and it will say BLANK in the brief citation | ||
| + | |||
| + | If there is not a Journal it will give an error | ||
| + | |||
| + | If there is a Journal: | ||
| + | |||
| + | if there is not a mapping to the mapping file it will give an error | ||
| + | |||
| + | if there is a Journal and there is a mapping and there is Full journal with stripped spaces it will print: | ||
| + | |||
| + | it will print "Journal_URL\t\"$mappings{strippedjourfull}{$journal}\"" | ||
| + | |||
| + | e.g.: Journal_URL "PLoSBiology" | ||
| + | |||
| + | if there isn't a stripped Journal name it will give an error | ||
| + | |||
| + | It will do the same thing for the Publisher_URL | ||
| + | |||
| + | it will print: "Publisher_URL\t\"$mappings{strippedpubname}{$journal}\"\n" | ||
| + | |||
| + | Publisher_URL "PLoS" Daniela add stripped name as for Jounal_URL | ||
| + | |||
| + | if there is a Journal and there is a mapping and there is a URL_accession it will print: | ||
| + | |||
| + | if ($data{urlaccession}{$pgid}) { # new output line for Daniela 2011 02 22 | ||
| + | my ($urlaccession) = &filterAce($data{urlaccession}{$pgid}); | ||
| + | $entry .= "Article_URL\t\"$mappings{strippedjourfull}{$journal}\" \"id\" \"$urlaccession\"\n"; } | ||
| + | elsif ($urlacc{$wbpaper}) { | ||
| + | my ($urlaccession) = &filterAce($urlacc{$wbpaper}); | ||
| + | $entry .= "Article_URL\t\"$mappings{strippedjourfull}{$journal}\" \"id\" \"$urlaccession\"\n"; } | ||
| + | else { print ERR "$pgid no urlaccession for $wbpaper\n"; } | ||
| + | |||
| + | e.g.: Article_URL "PLoSBiology" "id" "0030053" | ||
| + | |||
| + | home/acedb/draciti/oa_picture_ace_dumper/dump_picture_ace.pl now reads /home/acedb/draciti/picture_source/picture_source If the file isn't there or can't be read there's an error message and the program stops. Data from the 3rd column (always the third column) gets associated with data from the 1st column. If there's no data for the pg table pic_urlaccession, it looks to see if there's a match from the pic_reference table to the mapping on picture source, and uses the data from the third column as the urlaccession. Otherwise it gives an error. | ||
| + | |||
| + | |||
| + | If there isn't take the Accession number from the picture_source file (where there is mapping paper <-> Acc No) | ||
| + | if there is no Accession number in any of the 2 will give error | ||
| + | |||
| + | if there is a Journal and there is a mapping and there is a template it will print the template: | ||
| + | |||
| + | Template "WormBase thanks the journal <Journal_URL> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Journal_URL>, <Article_URL>. Copyright <Publication_year> with permission from <Publisher_URL>" | ||
| + | |||
| + | if not will give an error. | ||
| + | |||
| + | |||
| + | The following part of the dumper has been deleted after Paul Davis suggestions (feb 9th 2011) as the URL for the paper has been added to the Journal Database and the Brief citation will be pulled out directly from the ?Paper object by the webteam. | ||
| + | |||
| + | if there is a Journal and there is a mapping and there is a URL_accession it will print: | ||
| + | |||
| + | Article_URL\t${wbpaper}_URL id $urlaccession | ||
| + | |||
| + | e.g.: | ||
| + | |||
| + | Article_URL WBPaper00024505_URL id 0020352 | ||
| + | |||
| + | it will also creat the database object for the Article_URL | ||
| + | |||
| + | it will be | ||
| + | |||
| + | "Database : ${wbpaper}_URL" | ||
| + | |||
| + | If there is an Article_URL it will print | ||
| + | |||
| + | URL_constructor\t\"$mappings{arturl}{$journal}\" | ||
| + | |||
| + | my ($brief_citation) = &getBriefCitation( $firstauthor, $year, $journal, $title ); # from package /home/postgres/work/citace_upload/papers/get_brief_citation.pm | ||
| + | Name\t\"$brief_citation\" | ||
| + | |||
| + | There are "" that have to be escaped with backslash \ otherwise the .ace file is not reading in fine | ||
| + | |||
| + | e.g.: Database : WBPaper00024505_URL | ||
| + | Name "Gaudet J et al. (2004) PLoS Biol \"Whole-genome analysis of temporal gene expression during foregut ....\"" | ||
| + | URL_constructor "http:\/\/www.plosbiology.org\/article\/info:doi%2F10.1371%2Fjournal.pbio.%S" | ||
| + | |||
| + | ==Symbols conversion in the dumper script== | ||
| + | While dumping the following symbols will be converted: | ||
| + | |||
| + | µ in u | ||
| + | |||
| + | ± in +- '''change it in +/-''' | ||
| + | |||
| + | " are escaped in the following fields: description, source, remark, and urlaccession | ||
| + | |||
| + | multiple spaces in the "Description" section will be converted in to one single space | ||
| + | |||
| + | μ converts in u | ||
| + | |||
| + | α converts in alfa '''change it in alpha''' | ||
| + | |||
| + | ¡C converts in C | ||
| + | |||
| + | ° converts in C | ||
| + | |||
| + | Ð converts in - | ||
| + | |||
| + | |||
| + | ′ converts in ' | ||
| + | |||
| + | ¼ converts in u | ||
| + | |||
| + | ± converts in alfa '''change it in alpha''' | ||
| + | |||
| + | âÂÂ1 converts in - | ||
| + | |||
| + | > converts in > | ||
| + | |||
| + | < converts in < | ||
| + | |||
| + | â² converts in ' | ||
| + | |||
| + | β converts in Beta | ||
| + | |||
| + | |||
| + | '''âÂÂ¥ converts in ≥''' | ||
| + | |||
| + | '''∼ converts in ~''' | ||
| + | |||
| + | '''âü converts in -''' | ||
| + | |||
| + | '''² converts in Beta (e.g. pgid 7493)''' | ||
| + | |||
| + | '''Ï€ converts in ∏''' | ||
| + | |||
| + | '''⬠converts in ∏''' | ||
| + | |||
| + | '''± converts in alpha''' | ||
| + | |||
| + | '''¼ converts in u''' | ||
| + | |||
| + | '''â converts in Delta''' | ||
| + | |||
| + | '''â⬲ converts in '''' | ||
| + | |||
| + | '''ââ°¥ converts in ≥''' | ||
| + | |||
| + | '''Ãâ converts in x''' | ||
| + | |||
| + | '''0Ãâ converts in x''' | ||
| + | |||
| + | '''° converts in °''' | ||
| + | |||
| + | '''â˼ converts in -''' | ||
| + | |||
| + | '''â⬲ converts in '''' | ||
| + | |||
| + | '''° converts in °''' | ||
| + | |||
| + | '''â⬲ converts in '''' | ||
| + | |||
| + | '''¼ converts in u I think we put this already but dumps it wrong ((e.g. pgid 7528 7659 and several more)''' | ||
| + | |||
| + | ==Web display== | ||
| + | |||
| + | Adrian's code has already accounted for the contact. If there is both a paper reference and a contact, both will be displayed on the paper page. When there is no reference and there IS a contact, then on the expression pattern page, the contact will be acknowledged by "Courtesy of <Person>". A | ||
| + | |||
| + | ==Notes== | ||
| + | |||
| + | J: I forgot to point that when doing the WBPicture object creation, if | ||
| + | you need to edit the name of a picture object, you should be careful | ||
| + | not to have extra spaces around it or extra / missing digits, and that | ||
| + | the uppercase / lowercase is all okay. If you associated something as | ||
| + | being cropped to WBPicture0000000001 (or later under Expr_pattern), | ||
| + | and you then want to change it to WBPicture0000000002 don't forget | ||
| + | that you changing the object ID will _not_ change all associations to | ||
| + | it, so if picture 5 is still associated with picture 1, you'll still | ||
| + | have to query picture 5 to change its associated picture 1 to a | ||
| + | picture 2. and also it's an ontology, so if picture 1 is no longer a | ||
| + | valid picture, it might not come up in the OA to change, so you'd have | ||
| + | to delete all associations to picture 1 first. then change 1 to 2. | ||
| + | then reassign those associations to picture 2. | ||
| + | |||
| + | I imagine this will almost never be a problem for picture objects | ||
| + | (like it is for genes being merged and split), but you should be aware | ||
| + | of it. (and probably ask about if it's not clear, and put it on the | ||
| + | wiki in some section about renaming objects or something like that). | ||
| + | If you change the name right after duplicate, you wouldn't have | ||
| + | associated anything to it, so it would be okay. And I imagine that's | ||
| + | what you'll mostly be doing, so it should be okay. | ||
| + | |||
| + | ==Draft OA for picture curation== | ||
| + | |||
| + | WBPicture "" // this will be the picture ID -> generates automatically upon entry. We should have a "duplicate" button which generates a new ID. The object ID for the name reflects the postgres ID (pgid). Actually, the way the code is laid out, duplicate cannot assign a new pictureID, it has to duplicate the existing object ID. OK, no problem, I will do as Karen does! I was thinking of the way that date_last_updated changes in the GO config, but even then for duplicates it duplicates the old date, sorry =( But if the picture ID will always be the postgres ID, you can change the number based on the number in the pgid field. When Karen creates a new molecule object (only other config that creates IDs automatically), she still has to change the name too. What should the IDs look like ? It should be WBPicture0000000001 and progressive numbers --D Ok, changed from WBPicture:12345678 to WBPicture1234567890 (no : and 10 digits) Daniela has double checked with Gary -> Is OK to remove colon, which is used mainly for ontologies -- J J can you please change the Name into WBPicture in the OA? Done - J | ||
| + | |||
| + | Reference "" // This will record which paper this picture comes from -> ontology This is getting expr_pattern data from table obo_data_pic_exprpattern '''TODO change this when Expr Pattern OA is live'''. Make note on wiki for Expr Pattern OA -- J. OK J when I start the Expr_pattern OA wiki I'll make a note TODO Daniela--D Get the Jpgs from the picture_source file in Tazendra Daniela specify path for J. For now I want to be part of the automatic term info update as opposed to manual. I put the file "picture_source" on tazendra under draciti J please add Journal_name and Publication_year. And always show those 2 fields so that it is clear for me when is missing D TODO on tazendra create obo_ tables for pic_picturesource at /home/postgres/work/pgpopulation/obo_oa_ontologies/create_obo_pic_picturesource.pl -- J done 2010 12 21 Have moved picture_source file on mangolassi to /home/acedb/draciti/picture_source TODO on tazendra, incorporate to cronjob /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl -- J done 2010 12 21 Reference Term info now always has picture_source .jpg files listed, as well as Journal and Year (or BLANK for journal / year if not available) Rewrite /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl to use new format generated by mergeToOICR.pl script on Daniela's computer after Daniela makes it clear how to deal with .txt or .docx or what-not. -- J Juancarlos I have converted all the .docx files into .txt files. I would like to see a link to the .txt file in the term info (same place where I see the file names). If I understood correctly the mergeToOICR script should be changed including the .txt and then I will do the same as before. Scp to Tazendra and rsync with Canopus I've updated the script on canopus, so that it should work if you copy it to your computer (replace the one there, but maybe keep a copy just as a backup), and copied the new picture_source to mangolassi and populated the term info. But there are no WBPerson entries, so I can't test if that's going to work in the Term Info for contact. -- J I Copied the script on my computer, no problem that we cannot test the WBPerson Term Info yet. everything seems to work fine D We should add the locus information displayed together with the Expr_pattern in the Paper term info display. To do that: For every expr_pattern, look at the expr_pattern ontology to find Gene Name then use same mapping as GO OA to map gene to locus. If there is no locus put synonym. Thanks :) paper term info's expr now maps to a wbgene from obo_data_pic_exprpattern, and that maps to a locus from gin_synonyms gin_seqname gin_locus J | ||
| + | |||
| + | Update from Nov 23rd. D and J agreed that the pictures will be put on Canopus and J will take the picture source from there: /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl adapt this script to once a day crawl all the picture folders on canopus to get mapping of WBPapers to all their picture. Need to know what the picture folders are, and they need to be viewable from a webbrowser. | ||
| + | |||
| + | Contact "" // person multiontology on people as in Phenotype TODO update person term info to have contact from obo_data_pic_picturesource once Daniela tells me how to parse the file for .txt / .docx -- J. Same as for Reference D No data in Canopus with WBPerson, so can't test to see if it works -- J OK D Displaying term info in Contact field Fix script to populate the obo_data so that it will display file names in term info (.jpg and .txt)D | ||
| + | |||
| + | Description "" // this will be figure legend -> big text | ||
| + | |||
| + | Source "" // this will be the actual picture name -> small text J, when you are in the office, I will show you a file I might use for autocomplete the source. i don't know if it is feasible but we should have a look at it cause it can save me lots of copy-pasting ^^ If it is not ok we keep the small text field D okay, but if you want a set of stuff to autocomplete, you'd have to maintain something for the database to update from it. for example, there's a lot of obo files maintained in cvs in sourceforge, so there's a script that updates based on those. if you wanted to commit your file to some cvs repository like that, it should work -- J. OK once you see the file in the office you can tell me how easy it is to maintain that file -- D Ok, sure. It's more an issue of where you're going to keep it for a script to pick it up - J Daniela todo -> scp on Tazendra a file the file Picture_source and update it constantly every time you are done with a journal J I scp in tazendra under draciti a file called picture_source. D Oh, sorry, when we're live it should be on tazendra, for now since we're working on stuff I'm putting it on mangolassi. So you wanted to run a script manually to populate this (extra step) or did you want a cronjob to pick it up and update everyday (potential for 24 hour delay before you can curate, when would you want it to run ?). To be clear, what should I do with this file ? J If I would like to have a cronjob (update every day). If I understand correctly I will modify the file once in a while (whenever I have new data to put in) and the cronjob will automatically update the tables. If that is the case, let's go for it! Automatic, yes, but not instantaneous, I just want to be clear that if the script runs at 2am every day, and you update it at noon, you won't get to see the term info updates until the next day at 2am -- J No problem D. For Reference field, only display the JPGs when entering a paper. J you mean displaying only the JPGs coming from the picture_source file and not displaying the .docx files right? if this is what you mean the answer is yes. And if I am correct, in the Reference field, I will still continue to see expression pattern data, correct? D Yes, sorry, I meant just the filename of the jpg files, as opposed to the other files, and yes, in addition to other WBPaper data -- J OK D For source it doesn't matter because it's text, not ontology ? yes D For cropped_from, ignore it because it will autocomplete from this OA Source field, not from the picture_source information we get from this file ? Is all that correct ? -- J I think so.. Let's talk about this after the meeting to make sure, then you can confirm on this wiki -- J OK D now that I am a bit more free from the modelling I can finally seriously testing the OA. And regarding this, this morning is not working, maybe because you are working on it? ^^ The error I get is JSON parse failed D Sorry about that ! I had to wipe and repopulate the database on the sandbox for some interaction stuff a few times, and I forgot to recreate the picture tables afterwards (also, sorry, any data you entered is gone) - J No problem D | ||
| + | |||
| + | Cropped_from "" this will only be used by the cropped images to indicate its mother picture -> ontology of picture objects Single ontology. Have not done this yet since there are no real picture objects yet. What do you want to show in term info here? The cropped from will be used only for duplicated objects. Let's say I have a mother picture and I want to duplicate it because I have a cropped panel. I would like to see Cropped_from "journal.pbio.0020352.g006" You actually want the WBPicture ID here, right ? -- J. I want to have the same name as "source" of the mother pictureD What should it autocomplete on? It should autocomplete on the "source" of the mother picture --D Do you want anything on the term info, just the name and ID ? autocomplete only on source, not both source and ID ? If you can autocomplete on both source and ID would be good!D Sure (do reply to the PictureID stored in postgres, I'm pretty sure that's what you want, but do confirm. OK Juancarlos, maybe I confused myself. To summarize the Cropped_from field: In the Cropped_from field I want to autocomplete with the "source" of the mother picture. In the Term info I would like to see The name and the ID (e.g. WBPicture0000000001, and journal.pbio.0020352.g006). Does that sound right to you? otherwise I'll show you on Thursday -- D It kind of makes sense, but I'm not sure it's good. Each picture object has a picture ID, so when referring to it, it's best to refer to the picture ID, because it's potentially possible that the name could change. Otherwise we'd just have picture names instead of picture IDs, right ? So I think of the source as a name, and we could have this field autocomplete on the source/name, but then store/save the picture ID. Then when dumping to .ace outputting the source of that picture object. This way if you make ID 1 -> source "blah", then ID 2 -> cropped from ID 1, then change ID 1 -> source "different", when you dump picture 2 it would say cropped from picture ID 1, with source "different". If in the same case you put in picture 2 source -> "blah", when you dumped picture 2 it would always say "blah". Does that make sense ? -- J In the Cropped_from we will autocomplete on Source (file name) then WBPicture ID; and store in postgres the picture ID -- D Right, and we'll do that based on the Source OA field / postgres table, not what was entered in the "picture_source" file above -- J Yes! D Autocomplete on source then WBPicture name/ID. Show on Term Info name/ID, source, reference. -- J | ||
| + | |||
| + | Expr_pattern "" // this relates to the Expr_pattern associated with the picture -> multiontology File that has Expr_pattern <-> paper association is ExprWS221.ace received from Wen October, 22 2010. In term info we'd like to see Gene, Pattern, Reference, Reporter_gene, Life_stage Anatomy_term, GO_term. Autocomplete just on Expr_pattern ID. For Anatomy_term retrieve Anatomy_term ID and name from app_anatomy. Created obo_<data|name>_pic_exprpattern tables temporarily until Expr_pattern OA is live. TODO get rid of these tables once Expr_pattern OA is live. See below Make note on wiki for Expr Pattern OA. Created in mangolassi at /home/postgres/work/pgpopulation/exp_exprpattern/ When live on tazendra, TODO use /home/postgres/work/pgpopulation/exp_exprpattern/create_obo_pic_exprpattern.pl and populate_obo_exprpattern.pl DONE created on tazendra for grg_generegulation, which needed these tables. Also, since you requested the Expr_pattern field to be an ontology field, you should make sure that it works for you and Xiaodong's data, then tell her that if she wants it to work like that in her OA, she needs to let me know and then we need to transfer her data from text to ontology or multiontology. OK< Xiaodong said it is fine with her --D | ||
| + | |||
| + | ===Expr_pattern term info=== | ||
| + | Juancarlos, here is what I want to see in the picture OA for Expression Pattern term info | ||
| + | |||
| + | * Expr id : exp_name -- autocomplete on name only | ||
| + | |||
| + | * Gene : exp_gene -- show WBGenename, locus and synonym as in expr Pattern OA. e.g.: Oh, but this is a multiontology. Do you want to see all this info for all these genes in separate blocks ? I don't mind how is displayed actually, if it is easier for you could also be: whichever you like, I just thought it was confusing the other way, if everything is one line per data, then it's more clear in that sense | ||
| + | |||
| + | "WBGene00022781, pmt-1, ZK622.3, phi-40" | ||
| + | "WBGene00017066, maco-1, D2092.5" | ||
| + | |||
| + | It will be clear from the context that synonym means the synonym for that ID only ? I imagine every gene that has a synonym should have a unique synonym that'd be nice, but some genes are synonyms of other loci. You can tell from the paragraph context, but if we're doing it one line per wbgene, it's probably clearer | ||
| + | |||
| + | id : WBGene00022781 | ||
| + | locus : pmt-1 | ||
| + | synonym : ZK622.3 | ||
| + | synonym : phi-40 | ||
| + | |||
| + | * Anatomy_term : exp_anatomy exp_qualifier exp_qualifiertext --display all data one data per line e.g.: so for exp_anatomy show the name and the ID in doublequotes, yes and the other two things repeated for each term in exp_anatomy. the repeated things were exp_qualifier and exp_qualifiertext. I changed the example so maybe is more clear. | ||
| + | Anatomy_term : "nerve ring is WBbt:0006749" Certain expressed in XYZ | ||
| + | Anatomy_term : "pharynx is WBbt:0003681" Partial expression detected in a subset of neurons | ||
| + | |||
| + | |||
| + | * GO_term : exp_goid -- show GO ID followed by the GO name (I checked in GO OA and what I want to see here is "name" , I don't know how it is called in the GO tables e.g. ok, I'll figure it out great | ||
| + | GO_term : "GO:0005737" Cytoplasm | ||
| + | |||
| + | * Subcellular Localization : exp_subcellloc | ||
| + | |||
| + | * Life_stage : exp_lifestage -- Convert the life stage IDs into names from the obo_name_lifestage e.g. "L3 larva" so name only, no ID correct | ||
| + | |||
| + | * Antibody_text : exp_antibodytext | ||
| + | |||
| + | * Reporter Gene : exp_reportergene | ||
| + | |||
| + | * In_Situ : exp_insitu | ||
| + | |||
| + | * RT_PCR : exp_rtpcr | ||
| + | |||
| + | * Northern : exp_northern | ||
| + | |||
| + | * Western : exp_western | ||
| + | |||
| + | * Antibody_info : exp_antibody so this is the multiontology field, and the data looks like : "[cgc3991]:apr-1_b","[cgc3991]:apr-1_a" is good to have them in one line | ||
| + | |||
| + | * Pattern : exp_pattern | ||
| + | |||
| + | * Transgene : exp_transgene data looks like "kyIs136","kyIs131","kyIs137","kyIs140" fine | ||
| + | |||
| + | * Reference : exp_paper data looks like "WBPaper00001926","WBPaper00001469" fine | ||
| + | |||
| + | No western ? good catch!! I missed it :p np =) | ||
| + | |||
| + | |||
| + | Remark "" // For other curator notes -> big text Remark should be dumped as text. Try to generate few data in the OA and to dump a .ace file. See what will be dumped as remark. Tried (nov 28 2010) it works fine. | ||
| + | |||
| + | Cellular_component "" // For sub-cellular localization -> multiontology of GO_Term like gop_goid. File that has Expr_pattern <-> GO_term association is ExprWS221.ace received from Wen October, 22 2010. After I copy wen's file on tazendra and tell you to parse it MAKE PAPERS TERM INFO DISPLAY EXPR_PATTERN | ||
| + | |||
| + | Anatomy_term "" // It will link the picture object directly to an Anatomy Object -> multiontology. Should work like app_anatomy File that has Expr_pattern <-> Anatomy_term association is ExprWS221.ace received from Wen October, 22 2010. File that has Anatomy_term <-> anatomy name association is http://obo.cvs.sourceforge.net/viewvc/obo/obo/ontology/anatomy/gross_anatomy/animal_gross_anatomy/worm/worm_anatomy/WBbt.obo | ||
| + | For example, Wen's file has Anatomy_term "WBbt:0004854" | ||
| + | and OBO file has WBbt:0004854 name: vm1 def: "Vulval muscle 1" | ||
| + | |||
| + | Article_URL_Accession "" // Text this is the unique id pointing to a paper URL | ||
| + | |||
| + | Person "" person multiontology on people as in Phenotype | ||
| + | |||
| + | Person_text "" free text small. Note on the .ace file for Person and Person_name: join all person objects's starndard names with commas, then comma, then person_name text. If there's no person_name text. join all person objects's standard names with commas, except for the last one, which is joined by "<comma> and ". | ||
| + | |||
| + | Life_stage "" multiontology autocomplete on ExprWS221.ace received from Wen October, 22 2010 (same file as others) --D I don't understand what you mean about the Expr file, but you can see a life stage field in the phenotype OA, and see if that works like you'd want -J Yes, it si perfect to have the life stage field like in the phenotype OA D Ok, We have a few new tables to make, so I'll wait until we're set with those and make them all at once (curation_status, lifestage, anything else ?) -- J Give me a couple of days to test it more and to solve the Acknowledgment issue. I did not hear from the webteam yet and I feel bad coming back and forth to you with new requests! :( That's okay, we can wait for the acknowledgements, but are those the only things you want for non-acknowledgements ? Just curation_status and lifestage ? No. Have created tables urlaccession person persontext lifestage nodump chris 2010 11 15 Please test OA -- J Tested D | ||
| + | |||
| + | Curator "" field, where do you want it ? -- J Where you put it it is totally fine :) -D | ||
| + | |||
| + | No dump "" Toggle J please can you add the no dump field? Thanks D | ||
| + | |||
| + | Chris "" Toggle | ||
| + | |||
| + | |||
| + | Acknowledgments ""// the acknowledgment field will have more than one tag | ||
| + | |||
| + | * Template Text Daniela created a table with Publisher/template text association -> Mappings.txt tab delimited file | ||
| + | * Publication_year J will get data from paper tables | ||
| + | * Article_URL Daniela created a table with Journal name/URL constructor -> Mappings.txt tab delimited file | ||
| + | * Journal_URL Daniela created a table with Journal name/Journal_URL -> Mappings.txt tab delimited file | ||
| + | * Publisher_URL Daniela created a table with Publisher/Publisher URL -> Mappings.txt tab delimited file | ||
| + | * Person_name will have 2 boxes in OA | ||
| + | |||
| + | |||
| + | where | ||
| + | |||
| + | "Template" // is the template sentence e.g. "WormBase wishes to thank the journal <Journal_name> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Journal_name>, <Article_URL>. Copyright (<Publication_year>) with permission from <Publisher_URL>." The template sentence will change accordingly to what publishers need but the tags populating it will always be the ones listed below Okay, it sounds like you don't want to store the template in the OA, you'll just tell me to hardcode in the dumper script that if it's a given Journal, use this template, if another some specific other template, and so forth ; so that if the template ever changes for a given journal we can just change the dumper script, correct ? - J Supercorrect! -D Also, you should probably look at the full list of Journal objects, because there are _tons_ (1404) due to minor differences in spelling and what-not. You probably can't map all those to a template, but I don't know. Maybe once we have a list of papers (do we already ?) we can see what journals exist for those given papers. J I have a list of 184 journals containing Expr_pattern data but I don't have yet a number for the template sentences because I am still working on getting copyright and permissions. It can be there will be 5 or 50, I just don't know yet - J Right on, got it -- J You also need to work out the issue of pubmed_final with Kimberly (see Journal_name section) Here's the postgres query for the 1404 journals SELECT DISTINCT(pap_journal) FROM pap_journal ; which you can see on the referenceform.cgi linked in the sitemap -- J OK I wrote to Kimberly, her answer is that if a paper has a PMID and is missing Journal or Year, let her know. If it doesn't have a PMID and is missing that information, I should feel free to fill it in using the paper editor. | ||
| + | |||
| + | "Journal_name" // we can retrieve it from the ?Paper data model (?Paper Reference Journal UNIQUE ?Text) Yes, but you need to tell me what to do if there's no Journal info for a given paper. I need to write something in the code of the dumper script to account for cases where there's no Journal. I see. If you're entering a paper you can see that there is or isn't a journal, but if the pubmed_final field is not set to final, then seeing a journal doesn't always mean that there will be a journal later. You should talk to Kimberly about this. - J I wrote to Kimberly and asked her how we should proceed on that. I'll let you know as soon as she gets back to me D Hopefully we can talk to her after the conference call tomorrow -- J see Kimberly answer above D J please get data from Paper tables and If the journal-name is empty write BLANK D | ||
| + | |||
| + | "Publication_year" we can retrieve it from the ?Paper data model (?Paper Reference Publication_date UNIQUE ?Text) Same as above -- J Same as above -- D J please get data from Paper tables and If the Publication_year is empty write BLANK D | ||
| + | |||
| + | "Article_URL" // this will contain the URL pointing to the paper citation. Do you store that in postgres, or can we generate this from a paper ID pointing to WormBase ? -- Daniela will create tables journal name -> Article_URL | ||
| + | |||
| + | "Journal_URL" // this will contain the URL pointing to the journal. Do you store that in postgres, or can we generate this from a paper ID pointing to WormBase ? -- Daniela will create tables journal name -> Journal_URL | ||
| + | |||
| + | "Publisher_URL" this will contain the URL pointing to the publisher's homepage. Are the mappings always the same that we can get based on the journal name ? -- J Daniela will generate tables Publisher -> publisher URL | ||
| + | |||
| + | == To go live on tazendra == | ||
| + | |||
| + | psql -e testdb < /home/postgres/work/pgpopulation/pic_picture/create_tables done 2010 12 21 | ||
| + | |||
| + | /home/postgres/work/pgpopulation/obo_oa_ontologies/create_obo_pic_picturesource.pl done 2010 12 21 | ||
| + | |||
| + | /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl done 2010 12 21 | ||
| + | |||
| + | /home/postgres/work/pgpopulation/pic_picture/parse_pictures.pl > logfile done 2010 12 21 | ||
| + | |||
| + | set to cronjob (everyday at 2am) 0 2 * * * /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl done 2010 12 21 | ||
| + | |||
| + | tazendra picture OA is now live, verify that everything works properly then set the issue as resolved 2010 12 21 | ||
| + | |||
| + | (probably manually edit some entries that weren't parsed properly) | ||
| + | |||
| + | == Bugs and Fixes == | ||
| + | J added Chronograms names in the Expression pattern field in OA for old picture objects -> March 22nd 2011 | ||
| + | |||
| + | ==Pipeline for picture handling Development== | ||
| + | |||
| + | - set up the picture files handling as we discussed on skype. I would like to have the files on my computer and rsync it to canopus. On my computer I have a folder named Canopus/Pictures which contains 3 directories: 200/, 600/ and OICR.(I put the same thing on canopus for you to see /Users/danielaraciti/Desktop/Canopus/Pictures) I would like that all files in the 600/ will be renamed having a _600 extension before the .jpg. E.g.in directory called WBPaper00024399 the file called journal.pbio.0020280.g007_A.jpg should be named journal.pbio.0020280.g007_A_600.jpg. | ||
| + | |||
| + | Then I would need to have the files from /600 and /200 and put them all together in a new folder called OICR (run script ./mergeToOICR.pl). The script takes files from 600/ and 200/ and moves them in Pictures/OICR. | ||
| + | Once you run it it generates a file that you should scp into tazendra /home/acedb/draciti/picture_source/picture_source | ||
| + | |||
| + | mergeToOICR.pl script takes from Pictures/ there is 200/ 600/ OICR/ From 200/ take all folders and move to OICR/ From 600/ loop through each directory, and for each WBPaper####/ rename all pictures replacing .jpg with _600.jpg and move to appropriate OICR/WBPaper####/ directory. Do a check that all pictures in OICR/WBPaper####/ have a normal, _200, and _600 version. Also writes file picture_source with mappings of WBPaper to source.jpg names for the script that populates the appropriate .obo tables. Daniela will scp it to the proper location (/home/acedb/draciti/) after running the mergeToOICR.pl -- J | ||
| + | |||
| + | Script is in: /Users/danielaraciti/Desktop/Canopus/Pictures/mergeToOICR.pl | ||
| + | |||
| + | |||
| + | |||
| + | Then we rsync Pictures/OICR/ to Canopus. | ||
| + | |||
| + | ''rsync --progress --delete-after -a /Users/danielaraciti/Desktop/Canopus/Pictures/OICR/ daniela@canopus.caltech.edu:OICR/'' | ||
| + | |||
| + | |||
| + | Once this is done the 200/ directory should be empty and the 600/ directory has a list of empty folders which should be discarded. The next time I have a batch of pictures I recreate 600/ and 200/ and rerun ./mergeToOICR.pl | ||
| + | |||
| + | The very last step of the process is to adapt the following script as you described and to add in term info the .txt files. J did it on Dec 6th We now have a link in Reference and Contact to the .txt files. | ||
| + | |||
| + | /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl adapt this script to once a day crawl all the picture folders on canopus to get mapping of WBPapers to all their picture. Need to know what the picture folders are, and they need to be viewable from a webbrowser. Done | ||
| + | |||
| + | ==Reading in existing picture objects== | ||
| + | |||
| + | mangolassi : /home/postgres/work/pgpopulation/pic_picture/parse_pictures.pl | ||
| + | |||
| + | J: To make sure it works, we need to read in the existing picture objects from acedb, so scp the source file with the .ace objects you want populated to tazendra, and write somewhere on the wiki, how I should map the .ace data to these OA tables. | ||
| + | |||
| + | D: The file with existing picture objects is in tazendra and is called citace220picture.ace | ||
| + | |||
| + | in the file you have only 2 info for a single object: | ||
| + | |||
| + | Picture : "29055F14H3.11_1.jpg" -> this should map to the "source" field in the OA | ||
| + | |||
| + | Expr_pattern "Expr7505" -> this should map to the "Expr_Pattern" field in the OA | ||
| + | |||
| + | we need to add | ||
| + | |||
| + | Picture ID for each of them WBPicture0000000001, WBPicture0000000002 and so on | ||
| + | |||
| + | Reference "WBPaper12345678" | ||
| + | |||
| + | Hm, I'm not following. The data only has those two thing (jpg and expr_pattern), so where does the other data come from ? | ||
| + | / | ||
| + | |||
| + | Sure sorry, I thought you could take it from the expr_pattern data. I put another .ace file on tazendra called ExprWS221.ace where there are the Expr_patterns <-> Paper IDs. If this does not make sense at all, just let me know :) What about the Expr_pattern objects that have multiple papers ? -- J There should be no case in which an Expr_pattern object has multiple papers associated D At a glance, Expr_pattern : "Expr12" | ||
| + | has Reference "WBPaper00001926" Reference "WBPaper00001469" I haven't programatically looked into it though -- J | ||
| + | J Please use WBPaper00001926 as reference for Expr12. done - J | ||
| + | Comment: We are using the latest reference whenever there are 2 references associated to a picture. There were only 2 cases: Expr12 and the large scale study mentioned below : there is double correspondence for 2 papers, WBPaper00006525 and WBPaper00031006, for one single expression pattern and this is happening because there are 2 large scale studies but is anyway kind of weird. I spoke with Wen (who curated the data) and Raymond and we came up with the following consensus: we should associate to all those objects the reference WBPaper00031006 because is the most recent paper. If the second paper is WBPaper00031006, that's now the only paper, seems okay -- J OK --D | ||
| + | |||
| + | Look at mangolassi : /home/postgres/work/pgpopulation/pic_picture/errors There are many Chronograms which have no mappings to WBPapers, and many Expr_pattern objects that map to two papers (the same pair for all of them) -- J J thanks for looking into this. I checked Acedb and talked to Wen, all the chronograms are coming from a large scale study: Dupuy D et al. (2007) Nat Biotechnol "Genome-scale analysis of in vivo spatiotemporal promoter activity in ...." WBPaper00029359 I dumped a .ace file for chronograms, if you want to have a look at the file is on Tazendra in draciti and is called Chronograms.ace. I'm now also comparing against that file Great, thanks! | ||
| + | |||
| + | After this keep a hold on existing picture objects as there are pictures from Flickr and there are still some issues I have to resolve with Todd and Raymond. Thanks! D What do you mean by keeping a hold on existing picture object ? I don't understand -- J I mean that we should not dump any .ace file and put it in acedb yet. We want to generate new data and see if the process works fine with the new model --D I won't be dumping any data, I'll just make the dumper, and it's up to you to put it in acedb when you want to, but keep in mind that all data that gets populated in postgres will be dumper unless it has the NO DUMP flag. -- J OK, I just wanted to make sure. I think we discussed already that all the existing picture objects should be flagged as No dump--D | ||
| + | |||
| + | The file still has errors for Expr without reference. You can see it on mangolassi. Thanks Juancarlos, if there is no reference leave the "reference" field blank. This issue raised another problem, of course... :) J We need a new field in the OA called "Contact" a person multiontology on people as in Phenotype. It should be between "Reference" and "Description" and in the final.ace file should be dumped as Contact "standard name from ?Person" e.g. Contact "Raymond Lee". Please let me know if this is clear. I have put the description in the "Draft OA for picture curation" chapter and in "Final .ace file should be dumped as". I thought you were going to create persons that were personal communications, or some such, after talking to Kimberly. Don't you need a WBPaperID to be part of the Name field and Article_URL ? There is no Author field. -- J Juancarlos, I thought this through, I checked the Paper Data Model and pondered the pros and cons of having one solution over the other. I also asked Raymond an advice on that. The cleanest way to go is to add in the ?Picture model a Contact tag and therefore adding a Contact tag in OA. I hope this does not cause you too many problems. We can talk on Skype if you wish to. I have manually checked the file with the Errors (Expr without reference). Once we have added the "Contact" tag in the OA we will populate it as described below. I know the picture OA procedural way was a bit more challenging because we were developing the model in parallel with the OA but we are really almost there and I am extremely grateful to you for everything! So to summarize what should be done: | ||
| + | |||
| + | - Create a new field in the OA called "Contact" // person multiontology on people as in Phenotype. It should be between "Reference" and "Description" and in the final.ace file should be dumped as Contact "WBPersonID" e.g. Contact "WBPerson12028". Done | ||
| + | |||
| + | - Populate all the Expr without reference. All the entries that gave error should have WBPerson266 (Ian Hope) in the "Contact" field except: | ||
| + | |||
| + | b0523_5_phx.jpeg Expr35, b0523_5_vul.jpeg Expr35 which should have WBPerson1232 (Lynch AS) | ||
| + | |||
| + | c07b52v.jpeg Expr83, c07b54ec.jpeg Expr85, c07b54la.jpeg Expr85 Which now have a reference WBPaper00002319 (at that time it was in press) | ||
| + | |||
| + | J: I thought there was always one Expr per picture, but that is not the case : | ||
| + | * c09f9_3_larv.jpeg has many expr Expr2060 Expr2006 | ||
| + | * Expr3072_3073.png has many expr Expr3072 Expr3073 | ||
| + | * f59b2_13_can.jpeg has many expr Expr19 Expr8 | ||
| + | * f59b2_13_head.jpeg has many expr Expr19 Expr8 | ||
| + | * zc84_3_all.jpeg has many expr Expr25 Expr26 | ||
| + | * zc84_3_vnc.jpeg has many expr Expr25 Expr26 | ||
| + | |||
| + | And Expr19 has a paper, but Expr8 does not. Expr25 has, Expr26 does not. | ||
| + | |||
| + | I have checked this. It can well be that there is more than one expr_ per picture. However I don't know why Expr19 has a paper and Expr8 does not. I can fix this manually when we are live on tazendra. Fixed. Expr 8 is associated with Hope IA (1991) Develoopment 'promoter trapping' in Cenorabditis Elegans. 110329 DR | ||
| + | |||
| + | Also, 2 entries have description, it's probably easier if you enter those descriptions manually later if you want them (when it's live). For that matter, maybe the script can just populate the ones above and you can also fix those manually when it's live, since there's only 6 pictures. I think the best is that I fix everything manually when it's live. Can you just write here the ids of the 2 that have description? D Sounds good. No, the pgids could change if we change things or who-knows-what when we go live on tazendra. Just look at the .ace file, search for that tag, then query those objects when it's live. -- J OK D. The 2 objects that have a descroption associated are Expr_3071jpg and Expr3072_3073.png. Those were deleted from OA-- see below 110329 DR | ||
| + | |||
| + | Wiped the tables and populated this data by calling /home/postgres/work/pgpopulation/pic_picture/parse_pictures.pl | ||
| + | |||
| + | J: So the OA will have a Person field, Person_text field, and Contact field, all of which refer to people? | ||
| + | D: Yes. the reason for that is that the dumper constructs a sentence for the acknowledgments taking the Person field and combining it with the Person_text field. Therefore in acedb the Person_name is a text. We need instead something separate from the acknowledgment which refers only to the contact author, in case somebody wants the original file. That is the reason of why having the Contact tag | ||
| + | |||
| + | J: How does the dumper change to deal with ?Picture objects without a Paper ? (and what if they have no paper and no contact) | ||
| + | D: If there is no Reference it stays blank. There will be no such case in which we have no paper and no contact | ||
| + | |||
| + | |||
| + | J: Why make contact a multiontology (vs. ontology) | ||
| + | D: Because there could be cases in which the Principal investigators will be 2 | ||
| + | |||
| + | J: And why dump the Standard_name instead of the ?Person objects | ||
| + | D: You are right we should dump the ?Person objects | ||
| + | |||
| + | J: There's still a lot of stuff that's bold from before.I did not change them back because I thought they are reminders for you. | ||
| + | D: the stuff that is bold from before I left it bold because I thought they are comments for yourself, such as: TODO on tazendra create obo_ tables for pic_picturesource at /home/postgres/work/pgpopulation/obo_oa_ontologies/create_obo_pic_picturesource.pl -- J Have moved picture_source file on mangolassi to /home/acedb/draciti/picture_source TODO on tazendra, incorporate to cronjob /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl -- J Reference Term info now always has picture_source .jpg files listed, as well as Journal and Year (or BLANK for journal / year if not available) | ||
| + | |||
| + | J: Please make a separate ticket about the 600 / 200, and put in a different section of the wiki, as it doesn't really have to do with reading in existing picture objects. | ||
| + | D: OK | ||
| + | |||
| + | Also, in the future, please copy files to mangolassi instead of tazendra if we're doing the development there -- J Will do :)--D | ||
| + | |||
| + | Do you want me to make up the IDs starting from 1 and going forward ? Yes ok -- J | ||
| + | |||
| + | Where do the Paper IDs come from ? see above | ||
| + | |||
| + | We don't need a year because that's in the paper object, right ? right | ||
| + | |||
| + | We also need a curator, would that be you or someone else ? See below | ||
| + | |||
| + | Any other field that we should fill in that is required ? See below, there was more about the topic it goes down till the To Do chapter:) thanks! | ||
| + | |||
| + | I will add description and anatomy once I'll get to annotate them. You mean you'll enter them through the OA when it's live, not that you'll enter them into acedb, nor into the .ace file, right ? -- J Yes, I will enter them through OA when it's live D | ||
| + | |||
| + | |||
| + | J: Ok, but everything needs an ID and a curator, right ? What should it be for those ? | ||
| + | |||
| + | D:For the curator we can get the information from the file citace220Picture_timeStamp.ace that i put on tazendra under draciti. I will probably be the curator of the picture objects once I have associated the anatomy terms. For most of those the curator is Wen, but for many it says acedb or citace, which are not valid curators. You can tell me to assign them all to Wen (if Wen's okay with it, for Interactions she told Xiaodong to use Xiaodong as the curator) or be more specific.. -- J J put me as curator whenever it says acedb or citace, I have to go through them anyway.. :) D Well, in that case, which timestamp should I key the curator off of ? It would really be easier to just assign curator as a rule, but if you want to match the timestamp, I'd need to know which, because there's only one curator for the whole entry, but there are three curators in the timestamp. One after the Picture ID, one after the Expr_pattern tag, and one after the Expr_pattern data. This is not a real entry, but if it said this, which one should be the curator ? J put always me as curator also for existing picture objects, in this way it will be easier - D | ||
| + | |||
| + | Picture : "1_#9f7.jpeg" -O "2001-09-10_21:07:30_acedb" | ||
| + | |||
| + | Expr_pattern -O "2002-04-01_18:48:26_citace" "Expr55" -O "2002-04-01_18:48:26_wen" | ||
| + | |||
| + | |||
| + | |||
| + | J:Anything else that should be assigned, like curation status or anything else ? -- J | ||
| + | |||
| + | D: At the moment they should also be flagged as No dump since I have to manually curate them | ||
| + | |||
| + | Thanks again J and please let me know if I missed something fundamental...;) can well be...:P Feliz fin de semana! Dani | ||
| + | |||
| + | Assign no_dump, curator, source, expr_pattern, reference, and that's it, correct ? -- J Correct And assign IDs starting from 1 and going forward WBPicture0000000001. Thanks!! D | ||
| + | |||
| + | ==== Files for old Picture_objects ==== | ||
| + | |||
| + | The total No of old picture objects belonging to the picture class is 7228. See citace220Picture.ace file in this dir /Users/danielaraciti/Desktop/Wormbase/Files from Wen/PictureObjects | ||
| + | |||
| + | |||
| + | We have deleted the no dump flag for all the old picture objects on March 22nd so that the old objects will be dumped but we did not update the history table. | ||
| + | |||
| + | main directory on canopus: /home/daniela/sort_old_pictures | ||
| + | |||
| + | Juancarlos put the old picture objects for sorting on canopus here: /home/daniela/sort_old_pictures/5367 | ||
| + | |||
| + | March 23rd. J converted the png files -coming from chronograms- into jpg files. Also the postgres tables were converted accordingly. The picture source file was converted in jpg. the same is true for the pictures having a jpeg extension. | ||
| + | |||
| + | convert file.png file.jpg | ||
| + | convert_failed_png_to_jpg.sh | ||
| + | convert_png_to_jpg.sh | ||
| + | |||
| + | J checked that all the pictures present in postgres till pgid 7228 (included) have a corresponding file on canopus. | ||
| + | |||
| + | the location on canopus for the old picture was originally: | ||
| + | |||
| + | 5367 picture files are stored in Canopus here: | ||
| + | daniela@canopus:/usr/local/wormbase/website-shared-files/html/images-website-classic/expression/patterns | ||
| + | |||
| + | 2233 pictures (related to chronograms) are in this dir | ||
| + | daniela@canopus:/usr/local/wormbase/website-shared-files/html/images-website-classic/expression/localizome$ | ||
| + | |||
| + | Adding up 5367 and 2233 we should have 7599 pics. Canopus has a total of 7595 pictures files -> 4 pics discrepancy could be other files | ||
| + | |||
| + | 3 files are on tazendra and NOT on Canopus. Check the list in compare_tazendra_to_canopus.outfile in canopus in the following dir: /home/daniela/sort_old_pictures | ||
| + | |||
| + | 10_#9f7.jpg in tazendra, not on canopus WBPaper00001469 | ||
| + | |||
| + | Expr3071.jpg in tazendra, not on canopus WBPaper00024532. This picture is a cartoon of the worm for the old renderings. It is also located on canopus on this location: /usr/local/wormbase/images_for_raymond/expression/assembled/Expr3071.png | ||
| + | |||
| + | Expr3072_3073.jpg in tazendra, not on canopus WBPaper00024532 | ||
| + | |||
| + | Expr3071.jpg and Expr3072_3073.jpg correspond to the publication WBPaper00024532. The pictures were annotated directly by Daniela form the original publication. The 2 entries for the old pictures (pgid 4460 and 4461) could be removed for 2 reasons: A- the 2 riginal jpg files are not on canopus (one of the 2 was retrieved in another location and happened to be a cartoon, see above) B- the annotation of the pictures coming from the paper is in picture objects 7459, 7460, 7461. pgid 4460 and 4461 removed from Postgres on March 24th | ||
| + | |||
| + | /usr/local/wormbase/images_for_raymond/expression/assembled/Expr3071.png | ||
| + | |||
| + | |||
| + | Acknowledgments for old picture objects were set by default to persons and not to papers, we had therefore removed the reference from the reference field and put it as brief citation in the remark field. This is because the standard rule for acknowledgment for entries that have papers will follow the paper pipeline and search for a mapping for the publication. We have fixed all those entries either manually or programmatically. The excel file with details on that is /Users/danielaraciti/Desktop/Wormbase/Old_Pictures. We had as well added the contact and Person name to all the entries till 7228 | ||
| + | |||
| + | For the old pictures named .png and .jpeg Juancarlos used Imagemagik to convert them in jpg in a way that all the picture file names are in jpg extension. This will make the downstream processing more uniform. All the chronograms were .png and were all converted in jpg. As well all the files named jpeg were converted in jpg. | ||
| + | directory : | ||
| + | /home/acedb/draciti/oa_picture_fixes/20110324_source_jpg/ | ||
| + | |||
| + | script : | ||
| + | fix_source_jpg.pl | ||
| + | |||
| + | ==== To Do ==== | ||
| + | Parse Expr.ace file into .obo file for term info and for paper term info. On tazendra at /home/acedb/draciti/Expr_pattern/ExprWS221.ace | ||
| + | |||
| + | We already did this for generegulation OA -- J | ||
| + | |||
| + | ==== Legend ==== | ||
| + | |||
| + | * text : text | ||
| + | * bigtext : like longtext, but makes the text box expand when you click in it so you can see everything you've written | ||
| + | * dropdown : few values | ||
| + | * ontology : controlled vocabulary (tell me where they come from) | ||
| + | * multiontology / multidropdown : (allows multiple values) | ||
| + | * toggle : on / off, yes/no etc. | ||
| + | |||
| + | ==Daniela TODO when live on Tazendra to fit in old Picture data== | ||
| + | |||
| + | * c09f9_3_larv.jpg (PGID5645) has many expr Expr2060 Expr2006 done 110329 DR | ||
| + | * Expr3072_3073.jpg has many expr Expr3072 Expr3073 Objects deleted --see above for details 110329 DR | ||
| + | * f59b2_13_can.jpg has many expr Expr19 Expr8 done 110329 DR | ||
| + | * f59b2_13_head.jpg has many expr Expr19 Expr8 done 110329 DR | ||
| + | * zc84_3_all.jpg (PGID7078) has many expr Expr25 Expr26 done 110329 DR | ||
| + | * zc84_3_vnc.jpg has many expr Expr25 Expr26 done 110329 DR | ||
| + | |||
| + | And Expr19 has a paper, but Expr8 does not. Expr25 has, Expr26 does not. done 110329 DR | ||
| + | Assign Manually both Expr_patterns to the pictures | ||
| + | |||
| + | Add Description for 2 entries: Expr3071.png and Expr3072_3073.png --see above for details 110329 DR | ||
| + | |||
| + | '''Another thing that would be good is to set up a chronjob for updating the Mappings.txt file.''' | ||
| + | |||
| + | ==Final .ace file should be dumped as== | ||
| + | |||
| + | Picture : WBPicture0000000001 | ||
| + | |||
| + | Description "Figure Legend: A. ..... B. ..... C. .... D ....."" | ||
| + | |||
| + | Name "WBPaper12345678_journal.pbio.0020352.g006_B" -- I need to have in this field the "<data from Reference>_<Data from Source>" | ||
| + | |||
| + | Cropped_from "journal.pbio.0020352.g006" | ||
| + | |||
| + | Remark "Some remark" | ||
| + | |||
| + | Expr_pattern "Expr1234" | ||
| + | |||
| + | Anatomy "WBbt:0005175" | ||
| + | |||
| + | Anatomy "WBbt:0003681" | ||
| + | |||
| + | Cellular_component "GO:123456" | ||
| + | |||
| + | Template "WormBase thanks the journal <Journal_URL> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Journal_URL>, <Article_URL>. Copyright <Publication_year> with permission from <Publisher_URL>." -- take this from Template Text from Mappings.txt | ||
| + | |||
| + | Publication_year "2004" | ||
| + | |||
| + | Article_URL WBPaper00024505_URL id 0020352 -- Reference_URL id accession number. | ||
| + | |||
| + | Journal_URL "PLoS Biology" -- Take Full Journal Name from the Mappings.txt file on tazendra | ||
| + | |||
| + | Publisher_URL "PLoS" -- Take it from Publisher_name from Mappings.txt file | ||
| + | |||
| + | Reference "WBPaper00024505" | ||
| + | |||
| + | Contact "WBPersonID" | ||
| + | |||
| + | |||
| + | |||
| + | Database : WBPaper00024505_URL -- Reference_URL | ||
| + | |||
| + | Name "Ding M et al. (2008) PLoS One \"The cell signaling adaptor protein EPS-8 is essential for C. elegans epidermal ....\"" -- take this from Brief citation from Paper model NB There are "" that have to be escaped with backslash \ otherwise the .ace file is not reading in fine. Brief_citation name coming from new module at /home/postgres/work/citace_upload/papers/get_brief_citation.pm -- J | ||
| + | |||
| + | |||
| + | URL_constructor "http:\/\/www.plosbiology.org\/article\/info:doi%2F10.1371%2Fjournal.pbio.%S" -- take this from Article_URL from Mappings.txt | ||
| + | |||
| + | |||
| + | |||
| + | Database : "PLoS Biology" Take it from Full Journal Name from Mappings.txt | ||
| + | |||
| + | Name "PLoS Biology" Take it from Full Journal Name from Mappings.txt | ||
| + | |||
| + | URL_constructor "http:\/\/www.plosbiology.org\/" -- take this from Journal_URL from Mappings.txt | ||
| + | |||
| + | |||
| + | |||
| + | Database : PLoS -- Publisher_name in Mappings.txt | ||
| + | |||
| + | Name "PLoS" -- Publisher_name in Mappings.txt | ||
| + | |||
| + | URL_constructor "http:\/\/www.plos.org\/" take this from Publisher_URL from Mappings.txt | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | .ace dumper at mangolassi at /home/acedb/draciti/oa_picture_ace_dumper/ | ||
| + | (actually at /home/postgres/work/citace_upload/picture/ and symlinked here) | ||
| + | |||
| + | called dump_picture_ace.pl | ||
| + | |||
| + | generates pictures.ace and pictures.err (errorfile, always look at this even if it's usually empty) -- J | ||
| + | |||
| + | |||
| + | Daniela, once you are in Mangolassi | ||
| + | |||
| + | cd acedb/draciti/oa_picture_ace_dumper | ||
| + | |||
| + | give command | ||
| + | |||
| + | ./dump_picture_ace.pl | ||
| + | |||
| + | it generates 2 files: picture.ace and pictures.err | ||
| + | |||
| + | ==Sample curation results for a parental image when parental ≠ cropped== | ||
| + | |||
| + | Name "WBPicture0000000001" | ||
| + | |||
| + | Reference "WBPaper00024505" | ||
| + | |||
| + | Descritpion "(A) A portion of the promoter sequence of K07C11.4 from C. elegans (bottom) aligned with its ortholog from C. briggsae (top). Boxed regions show conserved predicted PHA-4 binding sites and Early-1 and Early-2 elements. Site-directed mutations that disrupt Early-1 and Early-2 (“E2 + E1 Mut”) are shown below their respective wild-type (“E2 + E1 WT”) sequence from K07C11.4. | ||
| + | (B–E) Confocal images of mid-stage embryos expressing GFP under the control of the wild-type K07C11.4 promoter (B) or promoters with a mutation in Early-1 (C), Early-2 (D), or both Early-1 and Early-2 (E). Percentages are the fraction of transgenic embryos expressing GFP; the remainder of embryos do not express GFP. | ||
| + | (F) Expression of the wild-type K07C11.4 reporter in a subset of somatic gonad cells in an L4 animal (arrowheads). | ||
| + | (G) Mutation of the Early-1 element eliminates gonadal expression but does not strongly affect expression in other tissues, such as intestinal cells (arrows). | ||
| + | Dashed lines indicate the outline of the developing pharynx." | ||
| + | |||
| + | Source "journal.pbio.0020352.g006" | ||
| + | |||
| + | Expr_pattern "Expr3097" | ||
| + | |||
| + | Remark "N/A" | ||
| + | |||
| + | Cellular_component "N/A" | ||
| + | |||
| + | Anatomy_term "N/A" | ||
| + | |||
| + | Acknowledgments "WormBase wishes to thank the journal Genetics for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from Genetics, 166:151-60, Chen J, Li XJ, Greenwald I. sel-7, a positive regulator of lin-12 activity, encodes a novel nuclear protein in Caenorhabditis elegans. Copyright (2004) with permission from the Genetics Society of America." | ||
| + | |||
| + | ==Sample curation results for a daughter image== | ||
| + | |||
| + | Name "WBPicture0000000002" | ||
| + | |||
| + | Reference "WBPaper00024505" | ||
| + | |||
| + | Descritpion "Confocal images of mid-stage embryos expressing GFP under the control of the wild-type K07C11.4 promoter" | ||
| + | |||
| + | Source "journal.pbio.0020352.g006_B" | ||
| + | |||
| + | Cropped_from "journal.pbio.0020352.g006" | ||
| + | |||
| + | Expr_pattern "Expr3097" | ||
| + | |||
| + | Remark "N/A" | ||
| + | |||
| + | Cellular_component "GO_term" | ||
| + | |||
| + | Anatomy_term "WBbt:0003681 pharynx" | ||
| + | |||
| + | Acknowledgments "WormBase wishes to thank the journal Genetics for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from Genetics, 166:151-60, Chen J, Li XJ, Greenwald I. sel-7, a positive regulator of lin-12 activity, encodes a novel nuclear protein in Caenorhabditis elegans. Copyright (2004) with permission from the Genetics Society of America." | ||
| + | |||
| + | ==Sample curation results for a parental image when parental = cropped== | ||
| + | |||
| + | Name "WBPicture0000000003" | ||
| + | |||
| + | Reference "WBPaper00024876" | ||
| + | |||
| + | Descritpion "Expression Pattern of rom-1::nls::gfp | ||
| + | Expression pattern of the zhIs5[rom-1::nls::gfprom-1::] transcriptional reporter during vulval development. Images on the left (A, C, E, G, and I) show the corresponding Nomarski pictures with the arrows pointing at the Pn.p cell nuclei and the arrowhead indicating the position of the AC nucleus. | ||
| + | (B) A mid L2 larva before vulval induction with uniform rom-1::nls::gfp expression in all the Pn.p cells. | ||
| + | (D) An early L3 larva in which rom-1::nls::gfp expression was decreased in all VPCs except P6.p (see text for a quantification of the expression pattern). Note that the nuclei of hyp7 and the Pn.p cells that had fused to hyp7 displayed strong rom-1::nls::gfp expression (P1.p, P2.p, P3.p and P9.p in the example shown). | ||
| + | (F) A mid to late L3 larva in which P6.p had generated four descendants. Expression of rom-1::nls::gfp occurred only in the 3° descendants of P.4.p and P8.p after they fused to hyp7. | ||
| + | (H) An L4 larva during vulval invagination. No rom-1::nls::gfp was detectable in the 1° and 2° descendants of P5.p, P6.p, and P7.p, but the AC and the surrounding uterine cells displayed strong rom-1::nls::gfp expression. | ||
| + | (K) A late L2 to early L3 larva following the ablation of the precursors of the somatic gonad. No up-regulation of rom-1::nls::gfp in P5.p, P6.p, or P7.p was observed. The scale bar in (K) is 10 μm." | ||
| + | |||
| + | Source "journal.pbio.0020334.g003" | ||
| + | |||
| + | Expr_pattern "Expr3457" | ||
| + | |||
| + | Remark "N/A" | ||
| + | |||
| + | Cellular_component "WBbt:0004017 Cell" | ||
| + | |||
| + | Anatomy_term "N/A" | ||
| + | |||
| + | Acknowledgments "WormBase wishes to thank the journal Genetics for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from Genetics, 166:151-60, Chen J, Li XJ, Greenwald I. sel-7, a positive regulator of lin-12 activity, encodes a novel nuclear protein in Caenorhabditis elegans. Copyright (2004) with permission from the Genetics Society of America." | ||
| + | |||
| + | ==Model testing== | ||
| + | |||
| + | On the temrinal | ||
| + | |||
| + | Go to acedb_good folder | ||
| + | |||
| + | then | ||
| + | |||
| + | ./xace /Users/danielaraciti/Desktop/Wormbase/ACEDB/ts | ||
| + | |||
| + | you have opened the empty database ready for testing. | ||
| + | |||
| + | then you can -> EDIT -> Read models -> yes -> continue | ||
| + | |||
| + | |||
| + | |||
| + | NOTES | ||
| + | |||
| + | For testing the model, the path is reading the models.wrm file that is in ts -> wspec -> models.wrm | ||
| + | |||
| + | Note that all the files that go to acedb should be plain text, so if you want to modify the models.wrm file you should convert it into plain text (in text edit -> format make plain text) | ||
| + | |||
| + | when you test a new model you should first go into ts/wspec, replace the old model with the new one, save and launch again the ts | ||
| + | |||
| + | If you get an error that says error line No 123: | ||
| + | Edit -> Find -> Select line and you go to the selected line | ||
| + | |||
| + | At the moment the file I am playing around with is models.wrm. | ||
| + | the original file was named modelsoriginal.wrm | ||
| + | |||
| + | In case you need to add back Variation to the ?Picture data model add the following tag in ?Variation model: | ||
| + | Picture ?Picture XREF Variation #Evidence | ||
| + | |||
| + | and add in the ?Picture model the following Variation ?Variation XREF Picture | ||
| + | |||
| + | same true for RNAi and Transgene | ||
| + | |||
| + | |||
| + | CHANGES TO THE MODELS.WRM FILE | ||
| + | |||
| + | ?Picture Description Text // not modified | ||
| + | |||
| + | Name UNIQUE Text // Added in ?Picture | ||
| + | |||
| + | Crop Crop_picture ?Picture XREF Cropped_from // added in ?Picture | ||
| + | |||
| + | Cropped_from ?Picture XREF Crop_picture //added in ?Picture | ||
| + | |||
| + | Pick me to call Text Text // not modified | ||
| + | |||
| + | Expr_pattern ?Expr_pattern XREF Picture // not modified | ||
| + | |||
| + | RNAi ?RNAi XREF Picture // deleted from ?Picture and deleted the XREF to Picture in RNAi class | ||
| + | |||
| + | Variation ?Variation XREF Picture // deleted from ?Picture and deleted the XREF to Picture in Variation class | ||
| + | |||
| + | Transgene ?Transgene XREF Picture // deleted from ?Picture and deleted the XREF to Picture in Transgene class | ||
| + | |||
| + | Remark ?Text #Evidence // not modified | ||
| + | |||
| + | Cellular_component ?GO_term XREF Picture //added in ?Picture and added "Picture ?Picture XREF Cellular_component" in ?GO_term | ||
| + | |||
| + | Anatomy_term ?Anatomy_term XREF Picture //added in ?Picture and added Picture ?Picture XREF Anatomy_term in ?Anatomy_term | ||
| + | |||
| + | Acknowledgments Template Text // added in ?Picture | ||
| + | |||
| + | Journal_name Text // added in ?Picture | ||
| + | |||
| + | Publication_year Text // added in ?Picture | ||
| + | |||
| + | Article_URL ?Database ?Database_field ?Accession_number // added in ?Picture | ||
| + | |||
| + | Publisher_URL ?Database ?Database_field ?Accession_number // added in ?Picture | ||
| + | |||
| + | Person_name Text // added in ?Picture | ||
| + | |||
| + | Reference ?Paper XREF Picture // added in ?Picture and added Picture ?Picture XREF Reference in ?Paper | ||
| + | |||
| + | ==Delete Expression== | ||
| + | in the folder OA Picture fixes there is a list of picture objects that are parental images to which I asked J to mass delete the XREF to Expr_pattern in a way that the Expr_pattern page would not have linked the parental images. Location: | ||
| + | /home/acedb/draciti/oa_picture_fixes/20110321_delete_expression | ||
| + | |||
| + | file name: pic_exprpattern.pg | ||
| + | |||
| + | that's the backup of the 135 entries to delete. Done DR 14.04.2011 | ||
| + | |||
| + | go to http://tazendra.caltech.edu/~postgres/cgi-bin/referenceform.cgi | ||
| + | |||
| + | To check the list of parental images which have a cropped from (use this to see which picture object should have the Expr_pattern XREF removed) | ||
| + | SELECT * FROM pic_exprpattern WHERE pic_exprpattern IS NOT NULL AND joinkey IN (SELECT joinkey FROM pic_name WHERE pic_name IN (SELECT pic_croppedfrom FROM pic_croppedfrom) ); | ||
| + | |||
| + | ==Identifying and retrieving pictures associated with Expression Pattern == | ||
| + | |||
| + | * 1. Use the curation status form to get a list of papers that do have curatable images. This includes images for which a paper has been curated for gene expression and for which we hold copyright permission. | ||
| + | |||
| + | http://tazendra.caltech.edu/~postgres/cgi-bin/curation_status.cgi | ||
| + | |||
| + | select curator and click on Curation statistics option Page. Select Pictures. It will retrieve a list of papers that have been curated for Expression_pattern and could potentially have expression images. As of now (Aug 2013) it displays all the papers, please note that we might not have permission to reproduce images for all of them. | ||
| + | In order to get a list of papers that contain images and are curatable (for which we hold permission) one should | ||
| + | |||
| + | <pre> | ||
| + | |||
| + | In this directory on tazendra : | ||
| + | /home/acedb/draciti/picture_curatable/ | ||
| + | |||
| + | Journals foe which we have permission are in this file (add journals here if we obtain more): | ||
| + | journal_with_permission | ||
| + | |||
| + | And run this script : | ||
| + | picture_flagged_permission.pl | ||
| + | |||
| + | What it does is get the papers from this URL : | ||
| + | http://tazendra.caltech.edu/~postgres/cgi-bin/curation_status.cgi?action=listCurationStatisticsPapersPage&select_curator=two1&listDatatype=picture&method=any%20pos%20ncur&checkbox_cfp=on&checkbox_afp=on&checkbox_svm=on | ||
| + | |||
| + | and filter against the pap_journal table through the list of | ||
| + | journal_with_permission | ||
| + | |||
| + | You can run it like : | ||
| + | ./picture_flagged_permission.pl > out | ||
| + | |||
| + | And look at the file 'out' | ||
| + | |||
| + | </pre> | ||
| + | |||
| + | * 2. Go to Textpresso.dev and run the image extraction pipeline using the WBPaperID list gathered in step #1 --using Arun's script-- this is the location on Textpresso-dev: | ||
| + | |||
| + | /data1/Users/liyuling/Curator_related/expression_pattern/scripts/ | ||
| + | |||
| + | Command: ./02legends_images_pdftohtml.pl "pdf_dir" "output_dir" | ||
| + | And one more command to package the output_dir into tar.gz | ||
| + | tar -zcvf output_dir.tar.gz output_dir | ||
| + | |||
| + | * 3. Once you have the folders with the Pictures | ||
| + | |||
| + | |||
| + | Contacting publishers to request permission on a paper-by-paper basis is too time consuming. Along the lines of other mods (zfin) we will retrieve only images from journals that are open access or granted us blanket permission. We could revise this pipeline in a year and re-negotiate with major publishers (e.g. AAAS). | ||
| + | |||
| + | ==Flagging a paper negative for pictures == | ||
| + | * go to the curation status form: http://tazendra.caltech.edu/~postgres/cgi-bin/curation_status.cgi | ||
| + | * Select you name and click on 'Specific paper page' | ||
| + | * Enter the WBPaperID, e.g.: WBPaper00001234 | ||
| + | * Select Picture | ||
| + | * Hit 'Get results' | ||
| + | * In the 'New result' box select validated negative | ||
| + | |||
| + | ==Identifying and retrieving pictures associated with topics == | ||
| + | |||
| + | December 2014: All topic diagram pictures will be put in OA regardless of permission. We will set manually the no dump toggle for images that do not have permission. | ||
| + | the dumper will give an error message in case there is a topic for a WBpaper that does not have permission | ||
| + | |||
| + | *process objects only dump if there's a paper and the paper has a journal | ||
| + | *the script looks at a file listing papers with permission, which is located here: /home/acedb/draciti/picture_curatable/journal_with_permission | ||
| + | *all the rest of the pipeline is the same as for Expression images: gets Mappings Journal/Publisher from /home/acedb/draciti/oa_picture_ace_dumper/Mappings.txt and URL accession numbers from /home/acedb/draciti/picture_source/picture_source | ||
| + | |||
| + | --------------- | ||
| + | |||
| + | the following query will give you a list of the WBPapers flagged relevant for a specific topic. Modify only the WBbioprID | ||
| + | |||
| + | <pre>SELECT DISTINCT(pro_paper) FROM pro_paper WHERE joinkey IN (SELECT | ||
| + | joinkey FROM pro_process WHERE pro_process ~ 'WBbiopr:00000046') AND | ||
| + | joinkey IN (SELECT joinkey FROM pro_topicpaperstatus WHERE | ||
| + | pro_topicpaperstatus = 'relevant') ORDER BY pro_paper; | ||
| + | </pre> | ||
| + | |||
| + | In this directory on tazendra : | ||
| + | /home/acedb/draciti/picture_curatable/ | ||
| + | |||
| + | |||
| + | run like | ||
| + | ./picture_flagged_permission.pl file | ||
| + | |||
| + | to use | ||
| + | http://tazendra.caltech.edu/~acedb/draciti/papers_with_topic_pictures.txt | ||
| + | |||
| + | edit file at | ||
| + | /home/acedb/public_html/draciti/papers_with_topic_pictures.txt | ||
| + | |||
| + | the script will get the list of WBPaerIDs that are in the URL and compare it with the list of journals for which we have permission. We are using it to identify images of processes but it can be used for other classes as well. | ||
| + | |||
| + | NB: in the directory there are 2 files: | ||
| + | journal_with_permission | ||
| + | journal_with_permission_expr | ||
| + | |||
| + | *journal_with_permission lists the open access papers (the ones we consider curatable for topic) | ||
| + | if you run the script ./picture_flagged_permission.pl file | ||
| + | you will get a list of the papers curatable for topic pictures | ||
| + | |||
| + | *journal_with_permission_expr lists the papers for which we have blanket permission to display expression pattern images -including Elsevier | ||
| + | to get the list of papers you just run the script ./picture_flagged_permission.pl | ||
| + | |||
| + | whenever you need to add/delete a paper you just do it on the list and the changes will automatically reflect in OA | ||
| + | |||
| + | == Permissions == | ||
| + | Below is a list of Journals for which we hold blanket permissions or are open access. A more detailed list is on Lario/Publishers/Permissions in Curation status.xlsx | ||
| + | |||
| + | <pre> | ||
| + | Mol Biol Cell | ||
| + | Genome Biol | ||
| + | J Biol | ||
| + | Neural Dev | ||
| + | BMC Biol | ||
| + | BMC Cell Biol | ||
| + | BMC Dev Biol | ||
| + | BMC Evol Biol | ||
| + | BMC Genet | ||
| + | BMC Genomics | ||
| + | BMC Mol Biol | ||
| + | BMC Neurosci | ||
| + | BMC Physiol | ||
| + | Genetics | ||
| + | G3 | ||
| + | COMP FUNCT GENOM | ||
| + | PLoS Biol | ||
| + | PLoS Comput Biol | ||
| + | PLoS Genet | ||
| + | PLoS Pathog | ||
| + | PLoS One | ||
| + | J Cell Biol | ||
| + | J Gen Physiol | ||
| + | Biochem Biophys Res Commun | ||
| + | Biochim Biophys Acta | ||
| + | Cancer Cell | ||
| + | Cell | ||
| + | Cell Host & Microbe | ||
| + | Cell Metab | ||
| + | Cell Signal | ||
| + | Cell Stem Cell | ||
| + | Chemistry and Biology | ||
| + | Curr Biol | ||
| + | Dev Cell | ||
| + | Exp Cell Res | ||
| + | Gene | ||
| + | Gene Expr Patterns | ||
| + | Genomics | ||
| + | Immunity | ||
| + | J Mol Biol | ||
| + | J Struct Biol | ||
| + | Mech Dev | ||
| + | Mol Cell | ||
| + | Neuron | ||
| + | Structure | ||
| + | Trends Parasitol | ||
| + | </pre> | ||
| + | |||
| + | We obtained an 'ad hoc' permission for the following journals | ||
| + | |||
| + | <pre> | ||
| + | Biochem J | ||
| + | Biochemistry | ||
| + | Biol Cell | ||
| + | Biol Chem | ||
| + | Biotechniques | ||
| + | Comp Funct Genomics | ||
| + | DNA Cell Biol | ||
| + | Development | ||
| + | Dis Model Mech | ||
| + | Endocrinology | ||
| + | FASEB J | ||
| + | Folia Biol (Praha) | ||
| + | Genes Dev | ||
| + | Genome Res | ||
| + | J Am Soc Nephrol | ||
| + | J Biol Chem | ||
| + | J Cell Sci | ||
| + | J Exp Biol | ||
| + | J Histochem Cytochem | ||
| + | J Neurosci | ||
| + | PNAS | ||
| + | Proc Natl Acad Sci U S A | ||
| + | RNA | ||
| + | Sci Signal | ||
| + | Science | ||
| + | </pre> | ||
| + | |||
| + | ==Mappings for the new_fetch.pl script == | ||
| + | location of the script /home/acedb/draciti/picture_curatable/Pictures_next_level/new_fetch.pl | ||
| + | |||
| + | The script worked well for BMC, Genetics, JCell Biol but did not initially work for science direct journals. We troubleshooted and got it to work but then again science direct changed the url for the high quality picture files. This is not mantainable in the long run as they keep changing the urls so we have retrieved the images with Arun's script (for 240 papers). In the future we should probably just use Arun's script as it is more reliable and agnostic on url changes. | ||
| + | <pre> | ||
| + | Publisher Fetch_pictures journals | ||
| + | AMER SOC CELL BIOLOGY Mol_Biol_Cell/Molbiolcell Mol Biol Cell | ||
| + | BIOMED CENTRAL LTD BMC/BMC Genome Biol | ||
| + | BIOMED CENTRAL LTD BMC/BMC J Biol | ||
| + | BIOMED CENTRAL LTD BMC/BMC Neural Dev | ||
| + | BIOMED CENTRAL LTD BMC/BMC BMC Biol | ||
| + | BIOMED CENTRAL LTD BMC/BMC BMC Cell Biol | ||
| + | BIOMED CENTRAL LTD BMC/BMC BMC Dev Biol | ||
| + | BIOMED CENTRAL LTD BMC/BMC BMC Evol Biol | ||
| + | BIOMED CENTRAL LTD BMC/BMC BMC Genet | ||
| + | BIOMED CENTRAL LTD BMC/BMC BMC Genomics | ||
| + | BIOMED CENTRAL LTD BMC/BMC BMC Mol Biol | ||
| + | BIOMED CENTRAL LTD BMC/BMC BMC Neurosci | ||
| + | BIOMED CENTRAL LTD BMC/BMC BMC Physiol | ||
| + | GENETICS SOC AM Genetics/Genetics Genetics | ||
| + | GENETICS SOC AM Genetics/Genetics G3 | ||
| + | PLoS PLoS/PLoS PLoS Biol | ||
| + | PLoS PLoS/PLoS PLoS Comput Biol | ||
| + | PLoS PLoS/PLoS PLoS Genet | ||
| + | PLoS PLoS/PLoS PLoS Pathog | ||
| + | PLoS PLoS/PLoS PLoS One | ||
| + | ROCKEFELLER UNIV PRESS JCellBiol/JCellBiol J Cell Biol | ||
| + | Elsevier BiochemBiophysResCommun/BiochemBiophysResCommun Biochem Biophys Res Commun | ||
| + | Elsevier Cell/Cell Cell | ||
| + | Elsevier CurrBiol/CurrBiol Curr Biol | ||
| + | Elsevier DevCell/DevCell Dev Cell | ||
| + | Elsevier Gene/Gene Gene | ||
| + | Elsevier JMolBiol/JMolBiol J Mol Biol | ||
| + | Elsevier MechDev/MechDev Mech Dev | ||
| + | Elsevier MolCell/MolCell Mol Cell | ||
| + | Elsevier Neuron Neuron | ||
| + | |||
| + | |||
| + | |||
| + | For the following papers I will retrieve pics with Arun's script (too few to be worth a separate script) | ||
| + | HINDAWI PUBLISHING CORPORATION COMP FUNCT GENOM | ||
| + | ROCKEFELLER UNIV PRESS J Gen Physiol | ||
| + | Elsevier Biochim Biophys Acta | ||
| + | Elsevier Cancer Cell | ||
| + | Elsevier Cell Host & Microbe | ||
| + | Elsevier Cell Metab | ||
| + | Elsevier Cell Signal | ||
| + | Elsevier Cell Stem Cell | ||
| + | Elsevier Chemistry and Biology | ||
| + | Elsevier Exp Cell Res | ||
| + | Elsevier Gene Expr Patterns | ||
| + | Elsevier Genomics | ||
| + | Elsevier Immunity | ||
| + | Elsevier J Struct Biol | ||
| + | Elsevier Structure | ||
| + | Elsevier Trends Parasitol | ||
| + | |||
| + | |||
| + | </pre> | ||
| + | |||
| + | |||
| + | Correspondence with Publishers about Permission is on Lario/Publishers, arranged by alphabetical order | ||
| + | |||
| + | == Automating Publisher Permission request== | ||
| + | |||
| + | Total number of papers with attached Expr_pattern object in WS221: 2406. We have contacted 48 publishers in order to request permission for image display into Wormbase. We have been able to obtain permission to reproduce images for 1162 papers (26 major publishers) and we are negotiating with 7 publishers to obtain permission for additional 1182 papers (On May 12th 2011). 3 publishers either did not accept the request or asked for a fee to reproduce the figures (38 papers). 13 publishers did not get back (24 papers). | ||
| + | |||
| + | To obtain permission for newly published images we want to set up an automated system so that each image will be requested during the curation process. | ||
| + | We will set up a pilot with National Academy of Sciences since they agreed to receive requests as they come up. We will extend the system to other publishers once developed. | ||
| + | |||
| + | Pictures will be curated during normal curation process (e.g. with Expr_patterns). Once a month a cronjob will read into OA and send an e-mail to the publisher requesting permission. Sample e-mail: | ||
| + | |||
| + | <nowiki>Dear <Publisher_name> | ||
| + | |||
| + | we are requesting permission to reproduce the following material into Wormbase. | ||
| + | |||
| + | - volume number, issue number, and issue date | ||
| + | - article title | ||
| + | - authors' names | ||
| + | - Figure/table number | ||
| + | |||
| + | We will reproduce the material into Wormbase, a non profit educational website. | ||
| + | Intended audience: researchers and students. | ||
| + | Please click here if you are accepting to grant us permission or here if you do not agree. | ||
| + | |||
| + | Thank you for your collaboration. | ||
| + | |||
| + | |||
| + | Daniela Raciti, PhD | ||
| + | California Institute of Technology | ||
| + | Curator, www.Wormbase.org | ||
| + | Mailing Address:
Division of Biology
Mail Code 156-29
California Institute of Technology
1200 E. California Blvd. | ||
| + | Pasadena, CA 91125 | ||
| + | |||
| + | Office Phone: (626) 395-8613 | ||
| + |
E-mail: draciti@caltech.edu</nowiki> | ||
| + | |||
| + | |||
| + | If this set-up will turn out to be good, we will move on and contact 6 additional major publishers. In turn we can extend the pipeline to smaller publishers. | ||
| + | |||
| + | ==Fetch Pictures== | ||
| + | |||
| + | '''Contact Dev Biol again for permission (211 papers)''' | ||
| + | |||
| + | Juancarlos' script fetch_pictures.pl (/Users/danielaraciti/Desktop/Fetchpictures) retrieves images from jorunals. | ||
| + | So far, images were retrieved from: | ||
| + | * Development (The Company of Biologists) 281 papers (181 able to fetch, 100 required Arun's script) | ||
| + | * JBC (Amer Soc Biochemistry Molecular Biology) 109 papers (99 able to fetch, 10 required Arun's script) | ||
| + | * Genes Dev (COLD SPRING HARBOR LAB PRESS) 97 papers (70 able to fetch, 27 required Arun's script) | ||
| + | * Mol Biol Cell (AMER SOC CELL BIOLOGY) 104 papers (99 able to fetch, 5 required Arun's script) | ||
| + | * Journal of Cell Science (Company of Biologists) 55 papers (42 able to fetch, 13 required Arun's script) | ||
| + | * J Cell Biol (Rockefeller university press) 74 papers (65 able to fetch, 9 required Arun's script) | ||
| + | * Cell (Elsevier Cell Press) 104 papers (79 able to fetch, 25 required Arun's script) | ||
| + | * Neuron (Elsevier Cell Press) 58 papers (56 able to fetch, 2 required Arun's script) | ||
| + | * Biochem Biophys Res Commun (Elsevier) 33 papers (23 able to fetch, 10 required Arun's script) | ||
| + | * Gene (Elsevier) 23 papers (12 able to fetch, 6 required Arun's script, 5 missing) | ||
| + | * J Mol Biol (Elsevier) 46 papers (37 able to fetch, 9 required Arun's script) | ||
| + | * Mech Dev (Elsevier) 21 papers (19 able to fetch, 1 required Arun's script, 1 missing) | ||
| + | |||
| + | For the following journals (Elsevier publisher) we have received permission but the number of papers is too low for being worth to write a fetching script so we will retrieve figures using Arun's script (36 papers total) search done May 1st 2012: | ||
| + | |||
| + | * Biochim Biophys Acta | ||
| + | * Cancer Cell | ||
| + | * Cell Host & Microbe | ||
| + | * Cell Metab | ||
| + | * Cell Signal | ||
| + | * Cell Stem Cell | ||
| + | * Chemistry and Biology | ||
| + | * Exp Cell Res | ||
| + | * Gene Expr Patterns | ||
| + | * Genomics | ||
| + | * Immunity | ||
| + | * J Struct Biol | ||
| + | * Structure | ||
| + | * Trends Parasitol | ||
| + | |||
| + | For the following journals (Elsevier) we did not obtain permission but the number of papers is too low. Only FEBS Lett would be worth a try | ||
| + | *Arch Biochem Biophys 1 | ||
| + | *Biochimie 1 | ||
| + | *Brain Res 2 | ||
| + | *Mol Brain Res 1 | ||
| + | *Chemical Physics 1 | ||
| + | *DNA Repair (Amst) 5 | ||
| + | *Eur J Cell Biol 2 | ||
| + | *Exp Gerontol 1 | ||
| + | *FEBS Lett 19 | ||
| + | *Free Radic Biol Med 1 | ||
| + | *Int J Parasit 7 | ||
| + | *J Neurosci Methods 2 | ||
| + | *Mech Ageing Dev 6 | ||
| + | *Mol Biochem Parasitol 3 | ||
| + | *Mol Cell Biol Res Commun 1 | ||
| + | *Mol Cell Neurosci 2 | ||
| + | *Molecular & Biochemical Parasitology 1 | ||
| + | *Mutat Res 1 | ||
| + | *Neurobiol Aging 1 | ||
| + | *Neuropharmacology 1 | ||
| + | *Neurosci Lett 3 | ||
| + | *Neuroscience 1 | ||
| + | *Parkinsonism Relat Disord 1 | ||
| + | *Toxicol Lett 1 | ||
| + | |||
| + | ==Textpresso mining pipeline== | ||
| + | |||
| + | We are developing pipeline in order to achieve automatic picture retrieval. The first trial was done with Development journal as 281 papers contained gene expression pattern objects. Juancarlos wrote a script to fetch pictures directly from the journal web-page so that the quality of the pictures will be the highest possible. On lario: /Users/danielaraciti/Desktop/Fetchpictures/fetch_pictures.pl. the script is extracting also the url for each paper and that url will be automatically dumped in the .ace file without need to insert it in the OA. | ||
| + | |||
| + | Out of 281 papers, 181 returned a positive result and all the figures and figure legends were extracted. For the remaining 100 papers Arun set up a pipeline for figure and figure caption mining form the pdfs stored in Wormbase. Briefly, every figure in the pdf is extracted and associated with a figure legend in a way that the figure name figure legend name will correspond. Subsequently the pdf is scanned again and converted into html with a pdf to html converter and the figures are again extracted with a higher quality. The first round of extraction is not sufficient to achieve a decent quality. In addition, a rule for flagging positive matches was developed, in a way that every figure caption contains a positive or negative flag. | ||
| + | |||
| + | The set of 100 papers that went through Arun script were manually curated. 96 of them were taken for further analysis (1 paper did not contain pictures associated with Expr_pattern and I need to get back to the remaining 3 papers). The goal was to calculate the script recall and precision. | ||
| + | |||
| + | Recall = 119/143 = 0.832167832167832 | ||
| + | |||
| + | Precision = 119/193 = 0.616580310880829 | ||
| + | |||
| + | ==Importing Wormatlas dataset== | ||
| + | |||
| + | Wormatlas contains 1999 pictures of gene expression. They all come from the Moerman large expression study. Zeynep agreed to let us display the pictures in the picture page. Juancarlos and I queried postgres and we already had 1734 out of 1999 pictures. We have been able to download the remaining 265 (264 because one object was empty). | ||
| + | The 264 images don't have a direct link to an Expression pattern object. What we want to do is to create new picture object is to script the info that are in the pic name. e.g. first digits before the _ are correspnding to the locus (e.g. '''AH6.11'''_BC12595_GFP_a-2_1.jpg). In order to autopopulate the Expression Pattern field in the picture OA we cannot browse directly postgres tables because that large scale study had not been imported into OA (still on Citace minus). What we should do is to find the ExprID directly on ACeDB. Whenever there are multiple expression pattern objects corresponding to the same gene, we should pick the one related to this publication: Hunt-Newbury R et al. (2007) PLoS Biol. High-throughput in vivo analysis of gene expression in Caenorhabditis ... | ||
| + | |||
| + | Daniela and Juancarlos worked on getting the mapping sequence-name -> gene -> Expr-object to import them into OA. All the files are in the Wormatlas folder on tazendra /home/acedb/draciti/worm_atlas. | ||
| + | |||
| + | The parsing into Picture OA will be as follows: | ||
| + | |||
| + | *pgid from 9116 on | ||
| + | *pic_name from 9116 on | ||
| + | *pic_contact WBPerson427 | ||
| + | *pic_source name of the file e.g. B0024.14b_BC13961_GFP_a-1_1.jpg | ||
| + | *pic_exprpattern Exprxxxx | ||
| + | *pic_remark Hunt-Newbury R et al. (2007) PLoS Biol. High-throughput in vivo analysis of gene expression in Caenorhabditis ... | ||
| + | *pic_person WBPerson427 | ||
| + | *pic_curator WBPerson12028 | ||
| + | |||
| + | Script to populate: /home/postgres/work/pgpopulation/pic_picture/20110919_wormatlas/populate_wormatlas.pl | ||
| + | |||
| + | NB: sometimes one gene has associated more than one Expression pattern (see tazendra /home/acedb/draciti/worm_atlas/out2), e.g. picture 01E11.7_BC10244_GFPV_a-1_1bi.jpg, WBGene00006508 Expr6402,Expr6403. However in OA the picture is associated only to Expr6402 as the strain associated to 6402 is BC10244 while the strain associated to 6403 is BC14499. | ||
| + | |||
| + | |||
| + | ==Itai Yanai large scale import -WBPaper00041190== | ||
| + | |||
| + | In order to display pictures of expression time course we needed to generate expression objects. The objects (Expression and Picture) will be deleted once Wen will finish curating microarray for all species described in the paper and once we will have in place a way to generate images of expression on the fly - data will be retrieved directly from SPELL. | ||
| + | |||
| + | For now Daniela and Juancarlos have generated 2 .ace files, one for pictures and one for expression. the files are on CitaceMinus. The files are called expr_pattern_Yanai.ace and pictures_Yanai.ace | ||
| + | |||
| + | Expression pattern and Picture objects were given high numbers so when the new display system will be in place those could be deleted without affecting anything in OA. | ||
| + | |||
| + | Expression objects go from Expr1010178 to Expr1029229 | ||
| + | |||
| + | Picture objects go from WBPicture0001011201 to WBPicture0001030251 | ||
| + | |||
| + | there are 19052 objects for each class. | ||
| + | |||
| + | ==GeneAce old objects== | ||
| + | in May 2015 | ||
| + | *1000 more picture objects in WS compared to citace or citace minus (ftp://ftp.sanger.ac.uk/pub2/wormbase/STAFF/pad/Daniela/bad_pictures.ace) | ||
| + | *Paul Davis found them coming from GeneAce | ||
| + | *Have "pick_me_to_call" tag | ||
| + | *Represent images from Ian Hope's lab (old Sylvia Martinelli objects) | ||
| + | *Objects are redundant with what we already have and will be deleted | ||
| + | |||
| + | Daniela and Juancarlos working on checking redundancies and importing the descriptions form Sylvia Martinelli to the pic_description field in OA | ||
| + | files here: | ||
| + | /home/postgres/work/pgpopulation/pic_picture/20150528_compare_sylvia_data | ||
| + | |||
| + | matching lists the files that are in OA out of 1001 only 13 are not: | ||
| + | |||
| + | <pre> | ||
| + | NOMATCH UL#155C2.1.jpg | ||
| + | NOMATCH UL#155C2.2.jpg | ||
| + | NOMATCH UL#155C2.3.jpg | ||
| + | NOMATCH UL#64A1.1.jpg | ||
| + | NOMATCH UL#64A1.2.jpg | ||
| + | NOMATCH r107_1_emb.jpg | ||
| + | NOMATCH r107_1_gnp.jpg | ||
| + | NOMATCH r107_1_lar.jpg | ||
| + | NOMATCH r11a5.2_adult.jpg | ||
| + | NOMATCH r11a5.2_head.jpg | ||
| + | NOMATCH r11a5.2_tail.jpg | ||
| + | NOMATCH r11a5.2_tail2.jpg | ||
| + | NOMATCH r11a5.2_vulv.jpg | ||
| + | </pre> | ||
| + | Daniela checked them the original names (e.g. UL#155C2.1.jpg) were converted into UL#155C2_1.jpg -underscore instead of full stop- for file_name reasons. | ||
| + | We have them all but NOMATCH r107_1_emb.jpg | ||
| + | NOMATCH r107_1_gnp.jpg | ||
| + | NOMATCH r107_1_lar.jpg | ||
| + | |||
| + | |||
| + | the file out is the output of the descriptions that will go populate the Picture OA in the pic_description field | ||
| + | |||
| + | ==Obsolete Summary Pipeline for picture handling== | ||
| + | * Pictures are saved and organized in folders in Lario (Daniela's computer) as described above. The folder name will be the WBPaperID or the WBPersonID | ||
| + | * Generate 600/folder containing 600x600 full view with Photoshop image processor | ||
| + | * Generate 200/ folder containing original files and 200x200 thumbnails with Thumbs Up | ||
| + | * Go to /Users/danielaraciti/Desktop/Canopus/Pictures and run script ''./mergeToOICR.pl'' The script that juancarlos wrote will # get all subfolders from the 200/ folder, # move them to the OICR folder, # get all subfolders for the 600/ folder, # get each jpg in those subfolders, # for each of those create a new name for it that names has _600 in it before the .jpg | ||
| + | * Check for errors (should automatically display errors if any). 200/ will be empty and 600/ will contain a number of empty folders. Delete them. | ||
| + | * Go to /Users/danielaraciti/Desktop/Canopus/Pictures and scp the file that was generated - picture_source- to tazendra ''scp picture_source acedb@tazendra.caltech.edu:/home/acedb/draciti/picture_source/'' | ||
| + | * rsync the file to Canopus ''rsync --progress --delete-after -a /Users/danielaraciti/Desktop/Canopus/Pictures/OICR/ daniela@canopus.caltech.edu:OICR/'' | ||
| + | |||
| + | Now you should see the .jpg file names and a link to the .txt file for the figure legend in the term info (takes it from picture_source) and OICR should have access to Canopus to get the actual files | ||
| + | |||
| + | To update the term info display into picture OA on tazendra: | ||
| + | |||
| + | Go to | ||
| + | /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl | ||
| + | |||
| + | this script updates directly the term info display into the OA> You have now links to the new pictures. | ||
| + | |||
| + | |||
| + | OICR should take the pictures from: | ||
| + | |||
| + | canopus.caltech.edu:/usr/local/wormbase/pictures/picture_object | ||
| + | |||
| + | ===Obsolete Pipeline for picture handling=== | ||
| + | |||
| + | Picture are saved on Lario (draciti's computer) as JPG files in directories organized after the WBPaperID. The original files will be converted in 200x200 pixels thumbnails and 600x600 pixels full view according the following: | ||
| + | |||
| + | ====Generating 200x200px thumbnails==== | ||
| + | |||
| + | Thumbnails are generated using the freeware "ThumbsUp" (v4.4) ''a simple, drag-and-drop based utility to create thumbnails for a bunch of pictures and supports all image formats of Mac OS X and QuickTime (including PDF documents)''<ref>http://www.macupdate.com/info.php/id/11898/thumbsup</ref> | ||
| + | |||
| + | Trials for automation have been done with Photoshop (automated image processor) and MacOSX (Automator -> creation of Thumbnail images). With Photoshop automator is NOT possible to save the thumbnails in the same folder. | ||
| + | With MacOSX Automator is not possible to create thumbnails larger than 128px. | ||
| + | ThumbsUp allows generation of 200x200 in the same folder where the original files are. | ||
| + | |||
| + | Therefore, in this folder -called 200- we have the reference file and the 200x200 thumbnail | ||
| + | |||
| + | The file name for thumbnails is the same as the original picture with a _200 suffix | ||
| + | |||
| + | [[File:PictureN1.png]] | ||
| + | |||
| + | ====Generating 600x600px full view==== | ||
| + | |||
| + | 600x600 images are generated with photoshop (scripts -> image processor) and stored in a separate folder called 600. The architecture of the sub-folders is the same as the original. It is not possible to generate the 600x600 with Thumbs up because it will not maintain the folder architecture if saved in a separate folder. | ||
| + | |||
| + | ====Merging ref, 200x200, and 600x600 pictures==== | ||
| + | |||
| + | We need to programmatically rename all the pictures that are in draciti/600/ folder. Then we will take the files from /600 and /200 and put them all together in a new folder called Merged (run script ./put_stuff_together.pl - Daniela modify this with the correct name). The script takes files from 600/ and 200/ and puts it together in draciti/merged | ||
| + | |||
| + | Then we rsync draciti/merged to Canopus. | ||
| + | Once this is done I move 600/ and 200/ to draciti/done | ||
| + | The next time I have a batch of pictures I recreate 600/ and 200/ and rerun ./put_stuff_together.pl | ||
| + | |||
| + | |||
| + | As a general comment. For the beginning the pipeline for picture curation will be done publisher by publisher. Once a batch of pictures is downloaded from PLoS we will do a batch conversion. Later on when we will work with single papers we will try to make the picture conversion part of the curation. | ||
| + | |||
| + | |||
| + | |||
| + | ==Canopus scripts== | ||
| + | |||
| + | the script is called | ||
| + | transfer_from_Pictures_to_good.pl | ||
| + | |||
| + | and is located here on canopus | ||
| + | |||
| + | /home/daniela | ||
| + | |||
| + | the script transfers jpg files from $dirSource to $dirDest if subdirectory+file have been curated in picture OA. 2012 10 18. The script is | ||
| + | |||
| + | <pre> | ||
| + | use File::Copy; | ||
| + | </pre> | ||
| + | |||
| + | is a non standard module that needs to be installed | ||
| + | |||
| + | here is where pics are coming from | ||
| + | |||
| + | <pre> | ||
| + | my $dirSource = '/home/daniela/all_pictures'; | ||
| + | </pre> | ||
| + | |||
| + | this is where pics are going to | ||
| + | |||
| + | <pre> | ||
| + | my $dirDest = '/home/daniela/OICR/Pictures/'; | ||
| + | </pre> | ||
| + | |||
| + | |||
| + | this hash stores pics that have been curated. the structure of the %curated hash maps to the the paper or person, then the file | ||
| + | <pre> | ||
| + | my %curated; | ||
| + | </pre> | ||
| + | |||
| + | the following line requests to have the Yanai file | ||
| + | <pre> | ||
| + | my $yanai_file = 'pictures_Yanai.ace'; | ||
| + | </pre> | ||
| + | |||
| + | opens the Yanai file. Since Yanai's pics are not in OA we need to read from a flat file. Its capturing those and is mapping them to the curated Hash | ||
| + | |||
| + | *$curated{"WBPerson4037"}{$file}++ | ||
| + | *%curated -> WBPerson4037 -> filename | ||
| + | |||
| + | |||
| + | in the Yanai file the filename looks like this: | ||
| + | Name "WBGene00003442.jpg" | ||
| + | |||
| + | Then we are querying postgres for the picture source and the papers for the picture source | ||
| + | *SELECT pic_source.pic_source, pic_paper.pic_paper, pic_paper.joinkey FROM pic_source, pic_paper WHERE pic_source.joinkey = pic_paper.joinkey | ||
| + | |||
| + | we get the filename, paper, pgid and we put it in the curated hash %curated -> paper -> filename | ||
| + | |||
| + | <pre> | ||
| + | my ($filename, $paper, $pgid) = $row =~ m/<TD>(.*?)<\/TD>/g; | ||
| + | </pre> | ||
| + | |||
| + | |||
| + | now we are doing the same query again | ||
| + | |||
| + | *SELECT pic_source.pic_source, pic_person.pic_person, pic_person.joinkey FROM pic_source, pic_person WHERE pic_source.joinkey = pic_person.joinkey | ||
| + | |||
| + | but instead of querying for papers we query for persons | ||
| + | |||
| + | and doing the same things | ||
| + | |||
| + | we get the filename, paper, pgid and we put it in the curated hash %curated -> person -> filename | ||
| + | |||
| + | in this query there could be multiple people and this is the line doing it | ||
| + | |||
| + | <pre> | ||
| + | my (@persons) = $allpersons =~ m/(WBPerson\d+)/g; | ||
| + | </pre> | ||
| + | |||
| + | and it goes into | ||
| + | <pre> | ||
| + | %curated -> each_person -> filename | ||
| + | </pre> | ||
| + | |||
| + | <pre> | ||
| + | my %source; | ||
| + | my @source = <${dirSource}/*>; | ||
| + | foreach my $source (@source) { | ||
| + | if (-d $source) { | ||
| + | my (@subsource) = <${source}/*.jpg>; | ||
| + | my (@subsource2) = <${source}/*.JPG>; | ||
| + | foreach my $subsource (@subsource, @subsource2) { | ||
| + | if (-f $subsource) { | ||
| + | my (@stuff) = split/\//, $subsource; | ||
| + | my $file = pop @stuff; | ||
| + | my $dir = pop @stuff; | ||
| + | next unless ($curated{$dir}{$file}); | ||
| + | # print "SOURCE $file D $dir E\n"; | ||
| + | $source{$dir}{$file}++; } } } } | ||
| + | |||
| + | </pre> | ||
| + | |||
| + | it reads into the source dir, it looks at each thing that exists there. It only looks at directories. Opens the dir and looks for .jpg or .JPG and if any of those are files -as opposed to dir or symlinks- it gets the file and the dir and puts into curated directory file %curated -> directory -> file | ||
| + | and this works because the name of the directories are named WBPaper######## or WBPerson######## | ||
| + | |||
| + | the same thing happens for the destination hash for the destination source | ||
| + | |||
| + | |||
| + | <pre> | ||
| + | my %dest; | ||
| + | my @dest = <${dirDest}/*>; | ||
| + | foreach my $dest (@dest) { | ||
| + | if (-d $dest) { | ||
| + | my (@subdest) = <${dest}/*.jpg>; | ||
| + | my (@subdest2) = <${dest}/*.JPG>; | ||
| + | foreach my $subdest (@subdest, @subdest2) { | ||
| + | if (-f $subdest) { | ||
| + | my (@stuff) = split/\//, $subdest; | ||
| + | my $file = pop @stuff; | ||
| + | my $dir = pop @stuff; | ||
| + | $dest{$dir}{$file}++; } } } } | ||
| + | |||
| + | </pre> | ||
| + | |||
| + | |||
| + | <pre> | ||
| + | foreach my $dir (sort keys %source) { | ||
| + | unless ($dest{$dir}) { | ||
| + | my $newDir = $dirDest . '/' . $dir; | ||
| + | unless (-e $newDir) { | ||
| + | mkdir $newDir, 0755; | ||
| + | print "mkdir $newDir\n"; } } | ||
| + | foreach my $file (sort keys %{ $source{$dir} }) { | ||
| + | unless ($dest{$dir}{$file}) { | ||
| + | copy("${dirSource}/${dir}/$file", "${dirDest}/${dir}/$file"); | ||
| + | print "copy ${dirSource}/${dir}/$file ${dirDest}/${dir}/$file\n"; } | ||
| + | } # foreach my $file (sort keys %{ $source{$dir} }) | ||
| + | } # foreach my $dir (sort keys %source) | ||
| + | |||
| + | |||
| + | </pre> | ||
| + | |||
| + | if it exists in the source hash but it does not exist in the dest hash, it creates a directory and make writable-executable | ||
| + | It goes through each file and if it exists in the source but not in the destination is copying the file over | ||
| + | |||
| + | <pre> | ||
| + | foreach my $dir (sort keys %dest) { | ||
| + | my $deleteDir = 0; | ||
| + | unless ($source{$dir}) { $deleteDir = $dirDest . '/' . $dir; } | ||
| + | foreach my $file (sort keys %{ $dest{$dir} }) { | ||
| + | unless ($source{$dir}{$file}) { | ||
| + | unlink("${dirDest}/${dir}/$file"); | ||
| + | print "rm ${dirDest}/${dir}/$file\n"; } | ||
| + | } # foreach my $file (sort keys %{ $dest{$dir} }) | ||
| + | if ($deleteDir) { | ||
| + | rmdir $deleteDir; | ||
| + | print "rmdir $deleteDir\n"; } | ||
| + | } # foreach my $dir (sort keys %dest) | ||
| + | </pre> | ||
| + | |||
| + | If it exists in the destination but not in the source get rid of the files and delete them and deletes the directory | ||
| + | |||
| + | |||
| + | ==Virtual Worm images== | ||
| + | |||
| + | We will import Chris' renderings generated with Blender into a widget on the Anatomy pages. | ||
| + | |||
| + | The renderings are located here: | ||
| + | |||
| + | http://canopus.caltech.edu/virtualworm/Anatomy_Images/ | ||
| + | |||
| + | Juancarlos wrote a script to compare the names of the jpgs in the Virtual Worm directories above to the anatomy ontology obo file. | ||
| + | |||
| + | The script is on tazendra here: | ||
| + | |||
| + | cd azurebrd/work/parsings/daniela/20160624_virtualworm | ||
| + | |||
| + | output in 'out' | ||
| + | |||
| + | ==Obsolete PMID retrieval for pictures== | ||
| + | This is how papers containing expression data were retrieved for picture curation. This pipeline became obsolete after the curation status form went live ~Jan 2013 | ||
| + | |||
| + | *Secure shell to spica | ||
| + | *go to dir Daniela/GetPMID | ||
| + | *modify the file JournalList.txt with the name of the journals you want to retrieve the papers containing expression data | ||
| + | *run the script ./getJournalPMID.sh | ||
| + | *the output with the list of papers containing Expr dtata is in JournalPaperWithExpr.ace and contains the WBPaperID and the PMID | ||
| + | *transfer the JournalPaperWithExpr.ace to your local machine, rename it after the Journal name and put it in a folder called after the journal name. E.g. enerate a folder called Science and rename the .ace file science.ace. | ||
| + | *GREP the PMIDs identifiers e.g.: GREP PMID Science.ace > PMID. In this way you have a file containing all the PMIDs | ||
| + | *run Yuling script to generate a PMID list that could be copied in pubmed. Goto Desktop/Scripts/SCRIPT PMID and then run the command "./grab_pmid.pl input output" | ||
| + | *Transfer the output file into the "Science" folder | ||
| + | *Copy the PMID list and paste it in the Pubmed search box | ||
| + | *Click search and send to file choosing xml format. Save it in the "Science" folder as Science.xml | ||
Latest revision as of 16:44, 20 October 2023
links to relevant pages
Caltech documentation
Pictures
Contents
- 1 Picture Data Model
- 2 Picture Curation
- 3 Pipeline
- 4 Picture Data Model Proposal
- 5 Picture Data Model step by step explanation
- 6 Example
- 7 OA interface
- 8 TODO Daniela when generating Expr_pattern OA
- 9 mapping of OA fields to postgres tables
- 10 Test dumper script
- 11 Chronograms
- 12 .ace template for dumping
- 13 How the dumper works
- 14 Symbols conversion in the dumper script
- 15 Web display
- 16 Notes
- 17 Draft OA for picture curation
- 18 To go live on tazendra
- 19 Bugs and Fixes
- 20 Pipeline for picture handling Development
- 21 Reading in existing picture objects
- 22 Daniela TODO when live on Tazendra to fit in old Picture data
- 23 Final .ace file should be dumped as
- 24 Sample curation results for a parental image when parental ≠ cropped
- 25 Sample curation results for a daughter image
- 26 Sample curation results for a parental image when parental = cropped
- 27 Model testing
- 28 Delete Expression
- 29 Identifying and retrieving pictures associated with Expression Pattern
- 30 Flagging a paper negative for pictures
- 31 Identifying and retrieving pictures associated with topics
- 32 Permissions
- 33 Mappings for the new_fetch.pl script
- 34 Automating Publisher Permission request
- 35 Fetch Pictures
- 36 Textpresso mining pipeline
- 37 Importing Wormatlas dataset
- 38 Itai Yanai large scale import -WBPaper00041190
- 39 GeneAce old objects
- 40 Obsolete Summary Pipeline for picture handling
- 41 Canopus scripts
- 42 Virtual Worm images
- 43 Obsolete PMID retrieval for pictures
Picture Data Model
////////////////////////////////////////////////////////////////////////////////////
?Picture Description ?Text
Name UNIQUE Text
Crop Crop_picture ?Picture XREF Cropped_from
Cropped_from ?Picture XREF Crop_picture
Pick_me_to_call Text Text
Remark ?Text #Evidence
Depict Expr_pattern ?Expr_pattern XREF Picture
Anatomy ?Anatomy_term XREF Picture
Cellular_component ?GO_term XREF Picture
Acknowledgment Template UNIQUE Text
Publication_year UNIQUE Text
Article_URL UNIQUE ?Database UNIQUE ?Database_field UNIQUE ?Accession_number
Journal_URL UNIQUE ?Database
Publisher_URL UNIQUE ?Database
Person_name UNIQUE Text
Reference ?Paper XREF Picture
Contact ?Person
///////////////////////////////////////////////////////////////////////////////////
Picture Curation
The immediate goal of picture curation is to be able to obtain images of gene expression data from the literature and individual laboratories and display them in the WormBase gene expression page.
- We want display images related to the temporal or spatial (e.g., tissue, subcellular, etc.) localization of any gene in a wild-type background with different data types
- Reporter gene analysis
- Antibody staining
- In situ hybridization
- RT-PCR
- Western or Northern blot data
Pipeline
In the early phases of curation, pictures will be taken from open access journals (e.g. PLoS, BMC, Biomed Central LTD). During the process of open access image curation, other publishers will be contacted for obtaining copyright permissions.
The images should be saved and stored according to the following guidelines. The example shown below refers to a PLoS Biology paper but the rules of handling the pictures are universal and not "paper specific".
Overview
This is a mock page of the expression page for gene K07C11.4. We would like to see highlighted panel B and F with the figure capture describing the expression of the gene AND be able to access the original figure by clicking the "See original figure" button.
Downloading and saving the images
Pictures are downloaded in TIFF format from the original paper.
Pictures are saved with their original name in order to minimize editing from the curator. In this case the file is called “journal.pbio.0020352.g006”. The files are directly converted into JPEG. TIFF is not indicated as web display format. Avoid using special characters like ' * / in the file name.
The file is saved in a directory named after the WB paper ID. E.g.: WBPaper00024505, meaning that picture “journal.pbio.0020352.g006” has been downloaded from WBPaper00024505.
These 2 numbers together WBPaper00024505_journal.pbio.0020352.g006 will be UNIQUE IDENTIFIERS of the object, that we call Picture object 1 (WBPicture000000001). The ID WBPicture000000001 will be the NAME of the object (?Picture) in the Picture Data Model.
The path WBPaper00024505_journal.pbio.0020352.g006 will define the SOURCE of the object in the Picture Data Model.
Now look at the picture above: In our WormBase expression pattern page we don’t want to display the whole picture because it contains information not pertinent to the expression data. We therefore need to CROP the 2 pictures depicting expression of the gene in the Wild Type. We want to have only panel B and F.
Each panel is cropped from the original picture in Photoshop and the files are saved as “journal.pbio.0020352.g006_B” “journal.pbio.0020352.g006_F” in the same directory as before: WBPaper00024505
These will be respectively Picture object 2(WBPicture000000002) and Picture object 3 (WBPicture000000003).
To summarize till now:
Picture object 1: WBPicture000000001: WBPaper00024505_journal.pbio.0020352.g006
Picture object 2 WBPicture000000002: WBPaper00024505_journal.pbio.0020352.g006_B
Picture object 3: WBPicture000000003: WBPaper00024505_journal.pbio.0020352.g006_F
where WBPicture000000001 corresponds to the NAME of the object in the picture data model and WBPaper00024505_ journal.pbio.0020352.g006 corresponds to the SOURCE of the object in the Picture Data Model.
Question to web team: is it OK to keep the file names as proposed? -> Yes (Answer from TH october 6th)
At the same time, the text file associated with the entire figure WBPicture000000001, is saved with the same name as the figure -journal.pbio.0020352.g006- with a .txt extension. In this way we can make sure which figure legend goes with which picture.
Special case: what do I do when one single panel refers to multiple genes. E.g. In the example below, panel B displays the expression of 3 different genes. We will simply name the pictures Fig3_B1, Fig3_B2, Fig3_B3.
Let's go one step further...
Picture lineage
Picture object 1 is our PARENTAL IMAGE, we will display it only when the user will click on a “see original figure” link. Picture Objects 2 and 3 are our Daughter Images, which will be displayed on the gene expression page. See mock page below for a visual example:
We would like to keep the lineage relationship in order to know how images should be handled. In other words, we would like to know which image should be displayed in the expression pattern page and which should be displayed next to the "See original figure" link.
For that purpose, in the Picture Data Model we have the "Image lineage" tag.
There are cases in which parental image = daughter image. See picture below.
To the web team: in this case is the Picture Data Model proposed sufficient to determine that this picture should be displayed as PARENTAL or DAUGHTER? Yes
Picture size and format
All the pictures should be in JPEG format, if possible.
The picture size for thumbnails shown in the main gene expression page should be 200x200 pixels.
Picture size for the full view 600x600 pixels.
Picture size for the original file will be as big as needed.
NB: a note on 200x200 and 600x600 pixel size. This will not distort the pictures but just put a constraint on the maximum size of the thumbnail or the full image.
Summary Pipeline for picture handling
- Pictures are saved and organized in folders in Lario (Daniela's computer) as described above. The folder name will be the WBPaperID or the WBPersonID
For picture download there are 2 main pipelines:
- 1- fetch picture with the script fetchpictures.pl
- 2- fetch pictures via Arun's script
After downloading the pictures move them in the OICR folder (/Users/danielaraciti/Desktop/Canopus/Pictures/OICR).
- Run Juancarlos' script to generate the picture_source file: ./mergeToOICR.pl
- Go to /Users/danielaraciti/Desktop/Canopus/Pictures and scp the file that was generated - picture_source- to tazendra scp picture_source acedb@tazendra.caltech.edu:/home/acedb/draciti/picture_source/
- rsync the file to Canopus rsync --progress --delete-after -a /Users/danielaraciti/Desktop/Canopus/Pictures/OICR/ daniela@canopus.caltech.edu:OICR/
- Go to canopus and run the script /home/daniela/OICR/makethumb.sh. the script will now generate 200 and 600 thumbnails.
To update the term info display into picture OA on tazendra:
Go to /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl
this script updates directly the term info display into the OA> You have now links to the new pictures.
Canopus Accessibility:
this is the location from where OICR takes images and movies ftp://caltech.wormbase.org/pub/OICR/
on canoups every night, /home/daniela/OICR/* is copied over to /usr/local/wormbase/OICR/ and /srv/ftp/pub/OICR/ (where files are
accessible via ftp)
Example for movies
canopus.caltech.edu:/usr/local/wormbase/OICR/Movies
ftp://canopus.caltech.edu/pub/OICR/Movies
OICR file location:
/usr/local/wormbase/website-shared-file/html/img-static/movies
Naming folders after WBPersonID
When you have multiple contacts remember to call the folder where you store pictures with the WBPersonID of the first person inserted in the OA
Large Scale imports
When you import a large scale study remember to put only the contact and person and to put the publication in the remarks as a brief citation. This is to prevent conflicts in the picture fetching process from OICR. The pictures will be called after Paper OR Person.
Pictures added by other curators
a cronjob is checking daily if there are new objects added by other curators the cronjob is set on tazendra in the acedb account
- 0 1 * * * /home/acedb/draciti/cronjobs/daily_pictures_curator.pl
so every day at 1am
Picture Data Model Proposal
////////////////////////////////////////////////////////////////////////////////////
?Picture Description ?Text
Name UNIQUE Text
Crop Crop_picture ?Picture XREF Cropped_from
Cropped_from ?Picture XREF Crop_picture
Pick_me_to_call Text Text
Remark ?Text #Evidence
Depict Expr_pattern ?Expr_pattern XREF Picture
Anatomy ?Anatomy_term XREF Picture
Cellular_component ?GO_term XREF Picture
Acknowledgment Template UNIQUE Text
Publication_year UNIQUE Text
Article_URL UNIQUE ?Database UNIQUE ?Database_field UNIQUE ?Accession_number
Journal_URL UNIQUE ?Database
Publisher_URL UNIQUE ?Database
Person_name UNIQUE Text
Reference ?Paper XREF Picture
Contact ?Person
///////////////////////////////////////////////////////////////////////////////////
Picture Data Model step by step explanation
Picture Name of the picture object. E.g. WBPicture0000000001
Description Figure legend
Name For actual picture names. (This is the name of the path leading to the picture file. The source includes the name of the directory where the picture comes from AND the name of the picture file. e.g. WBPaper00024505_journal.pbio.0020352.g006. Deprecated). New decision made with web team. The "name" will only be the name of the file e.g.: journal.pbio.0020352.g006. The web team will construct the path as we discussed via e-mail on Dec 8th:
"So to back this up we will provide in the .ace file
Reference "WBPaper12345678"
Contact "WBPerson1234"
Name "pic.abcdefg.jpg"
and the rule to construct the path is:
knowing that if there is a reference the path will be Reference/Name (e.g. WBPaper12345678/pic.abcdefg.jpg) and if there is no Reference it will be Contact/Name (WBPerson1234/pic.abcdefg.jpg)" Daniela
Crop This is the picture object lineage. Large figures will be cropped into sections when they represent different data. We want to maintain the picture lineage -> by clicking on the "see original figure button" we want to access the entire image.
Pick_me_to_call Untouched tag from the existing model.
Remark For curator notes
Depict
Expr_pattern For linking to Expr-pattern data. This will be the Expr_pattern object that is associated with the picture.
Anatomy It will link the picture object directly to an Anatomy Object
Cellular_component This links to the GO term e.g. if a picture depicts sub-cellular localization
Reference
For the source of the picture E.g.WBPaper12345678.
Contact Whenever the picture does not come from a publication but from a person/lab this is the person who should be contacted. Normally the PI of the lab where the picture has been generated.
Note: the following tags were removed from the model:
RNAi
Variation
Transgene
because there were no data associated to those tags. The search to check association was done last time on November 4th on WS219
Acknowledgment
e.g.: WormBase thanks the journal Genetics <http://www.genetics.org/> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from Chen et al, Genetics 166:151-60, sel-7, a positive regulator of lin-12 activity, encodes a novel nuclear protein in Caenorhabditis elegans <http://www.genetics.org/cgi/content/full/166/1/151>. Copyright (2004) with permission from the Genetics Society of America <http://www.genetics-gsa.org/>.
In the sentence there are 4 variables:
"WormBase thanks the journal <Journal_URL> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Article_URL>. Copyright (<Publication_year>) with permission from <Publisher_URL>."
* Acknowledgment UNIQUE Template Text
* Publication_year UNIQUE Text
* Journal_URL UNIQUE ?Database
* Article_URL UNIQUE ?Database UNIQUE ?Database_field UNIQUE ?Accession_number
* Publisher_URL UNIQUE ?Database
* Person_name Text
where
Template Is the template sentence e.g. "WormBase thanks the journal <Journal_URL> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Article_URL>. Copyright (<Publication_year>) with permission from <Publisher_URL>."
The template sentence will change accordingly to what publishers need but the tags populating it will always be the ones listed below
Publication_year self explanatory
Journal_URL this will contain the URL pointing to the journal home page.
Article_URL this will contain the URL pointing to the paper citation.
Publisher_URL this will contain the URL pointing to the publisher's homepage.
Person_name if the picture is given by a person/lab
Update October 2023
- To allow consistency in links and to avoid broken links due to publisher's changes we did update the Article_URL so that it will point to the DOI. See this ticket for more information -https://github.com/WormBase/website/issues/9349.
Example
Picture : "WBPicture0000000001"
Description "Figure Legend: A. ..... B. ..... C. .... D .....""
Name "journal.pbio.0020352.g006_B"
Cropped_from "journal.pbio.0020352.g006"
Remark "Some remark"
Expr_pattern "Expr1234"
Anatomy "WBbt:0004017"
Cellular_component "GO:0005634"
Template "WormBase thanks the journal <Journal_URL> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Journal_URL>, <Article_URL>. Copyright <Publication_year> with permission from <Publisher_URL>."
Publication_year "2004"
Article_URL WBPaper00024505_URL id 0020352
Journal_URL "PLoSBiology"
Publisher_URL "PLoS"
Reference "WBPaper00024505"
Contact "WBPerson123"
Database : WBPaper00024505_URL
Name "Ding M et al. (2008) PLoS One \"The cell signaling adaptor protein EPS-8 is essential for C. elegans epidermal ....\""
URL_constructor "http:\/\/www.plosbiology.org\/article\/info:doi%2F10.1371%2Fjournal.pbio.%S"
Database : PLoSBiology
Name "PLoS Biology"
URL_constructor "http:\/\/www.plosbiology.org\/"
Database : PLoS
Name "PLoS"
URL_constructor "http:\/\/www.plos.org\/"
OA interface
Tab1
- Pgdbid - postgres database ID, generates automatically upon entry.
- WBPicture - Generates automatically upon entry. When duplicating a picture object be sure to assign a new Picture ID number which has the same number as the postgres ID (pgid). Be extra careful when you change name to an existing object. See section Renaming objects.
- Reference - WBPaperID paper ontology -
- Contact - Multiontology on people. N.B. when you have more than one Contact be aware on how to name the folder that stores pictures. As a general rule the folder should always be called after the first person inserted.
- Description - Figure legend (bigtext)
- Source - Multiontology from file picture_source on Tazendra (acedb@tazendra.caltech.edu:/home/acedb/draciti/)
- Cropped_From - Multiontology on source then WBPicture name/ID. Show on Term Info name/ID, source, reference.
- Expr_pattern - source file in Tazendra: /home/acedb/draciti/ExprWS221.ace. In term info we'd like to see Gene, Pattern, Reference, Reporter_gene, Life_stage Anatomy_term, GO_term. Autocomplete just on Expr_pattern ID.
- Topic - multiontology on process term
- Remark - bigtext
- Cellular component - multiontology of GO_Term like gop_goid. source file in Tazendra: /home/acedb/draciti/ExprWS221.ace.
- Anatomy term - multiontology. Should work like app_anatomy. source file in Tazendra: /home/acedb/draciti/ExprWS221.ace. File that has Anatomy_term <-> anatomy name association is https://github.com/raymond91125/Wao/raw/master/WBbt.obo. Previously was: http://brebiou.cshl.edu/viewcvs/*checkout*/Wao/WBbt.obo but became obsolete 04-11-2011 DR (and even before was http://obo.cvs.sourceforge.net/viewvc/obo/obo/ontology/anatomy/gross_anatomy/animal_gross_anatomy/worm/worm_anatomy/WBbt.obo but became obsolete 02-11-2011 DR)
- URL_Accession - text
- Person - Multiontology on people
- Person_text - free text. The person text was created for cases when we want to acknowledge people that are not WBPersons. Note on the .ace file for Person and Person_name: join all <pic_person> objects's starndard names with commas, then comma, then <pic_persontext> text. If there's no <pic_persontext>: join all person objects's standard names with commas, except for the last one, which is joined by "<comma> and ". Daniela remember for curation when you enter a text in the person text you should do the following: If it is a single entity in the text write e.g. "and Paul Sternberg's lab" if there are multiple entities e.g. summer student 1 (non WBPerson), summer student 2 (non WBPerson), and summer student 3 (non WBPerson) write "John Smith, George Brown, and Mike Lee". It is constructed in this way otherwise the syntax would have been too complicated
- Life_stage - like in the phenotype OA
- Curator - Multiontology on people
- No dump - Toggle
- Chris Flag - Toggle
*Permission - Dropdown with the following values: blank|Daniela|e-mail sent|granted|rejected
- Acknowledgment - the acknowledgment section is hard coded in the dumper script.
modify the dumper for acknowledgment for PNAS -not urgent, need to be ready when starting annotating PNAS papers. We need two different acknowledgments: till end of 2008 use this acknowledgment: Reprinted with permission from <Journal_URL> <Article_URL>. Copyright (<Publication_year>), <Publisher_URL>. From 2009 on use this: Reprinted with permission from <Journal_URL> <Article_URL>, <Publisher_URL>.
If there is a Reference the Acknowledgment is constructed this way: The information on Journal_name will be taken from Paper tables and If the Journal_name is empty write BLANK. The same is true for Publication_year, will be taken from Paper tables and If the Journal_name is empty write BLANK. The mapping file for the other fields of the acknowledgments is called Mapings.txt and is on Tazendra: /home/acedb/draciti/Mappings.txt. The file contains The template text, Article_URL, Journal_URL, and Publisher_URL.
When there is no Reference but there is a Contact the Acknowledgment will also be constructed automatically. Wormbase thanks <Person_name> for providing the pictures. The Person_name is constructed with the "Person" and "Person_text" tags (hardcoded in the dumper script). See details in the dumper section.
If a paper has a PMID and is missing Journal or Year, let Kimberly know. If it doesn't have a PMID and is missing that information, fill it in using the paper editor.
tab2
The postgres tables are
- Phenotype multiontology on phenotypes pic_phenotype
- alleleontology on variations pic_variation
- Gene multiontology on genes pic_wbgene
TODO Daniela when generating Expr_pattern OA
Other OA configs going to use WBPicture object ID: Expr_pattern OA (when it will exist) When coding complete: J set milestone to "code complete" D check that it works, set milestone to "verified" J make it live on tazendra, set milestone to "live" D check it works, and resolve the issue.
mapping of OA fields to postgres tables
- WBPicture -> pic_name
- Reference -> pic_paper
- Contact -> pic_contact
- Description -> pic_description
- Source -> pic_source
- Cropped_from -> pic_croppedfrom
- Expression Pattern -> pic_exprpattern
- Topic -> pic_process
- Remark -> pic_remark
- Cellular_component -> pic_goid
- Anatomy_term -> pic_anat_term
- URL Accession -> pic_urlaccession
- Person -> pic_person
- Person Text -> pic_persontext
- Life Stage -> pic_lifestage
- Species -> pic_species
- Curator -> pic_curator
- NO DUMP -> pic_nodump
- Chris Flag -> pic_chris
- Phenotype -> pic_phenotype
- Allele -> pic_variation
- Gene -> pic_wbgene
Test dumper script
On the Sandbox:
go to mangolassi
ssh acedb@mangolassi.caltech.edu
cd /home/acedb/draciti/oa_picture_ace_dumper
./dump_picture_ace.pl
the dumper generates 2 files: picture.ace and pictures.err
On Tazendra:
go to Tazendra
ssh acedb@tazendra.caltech.edu
cd /home/acedb/draciti/oa_picture_ace_dumper
./dump_picture_ace.pl
Chronograms
The chronogram could not be put in the Expr_pattern field. The Chronogram name will be put in directly frorm the dumper script. Juancarlos could not fit in Chronograms in the ontology as that applied only to Expr_patterns. In the .ace file we have anyway the tag "Expr_pattern "Chronogram1"" so the association in the picture page should display fine.
.ace template for dumping
Picture : <pic_name>
Description "<pic_description>"
Name "<pic_source>"
Cropped_from "<pic_croppedfrom>"
Remark "<pic_remark>"
Expr_pattern "<pic_exprpattern>"
Anatomy "<pic_anat_term>"
Cellular_component "<pic_goid>"
Template "<Template Text>" from Mappings.txt file on tazendra. When there is no Reference but there is a Contact it will generate automatically the template sentence Wormbase thanks <Person_name> for providing the pictures (hard-coded in the dumper).
Publication_year -- take this from Paper tables. Juancarlos I don't know the specifics of the Paper tables
Article_URL "<pic_paper>_URL id <pic_urlaccession>"
Journal_URL "<Full Journal Name>" from the Mappings.txt file on tazendra
Publisher_URL "<Publisher_name>" from Mappings.txt file on tazendra
Reference "<pic_paper>"
Contact "<pic_contact>"
Database : "<pic_paper>_URL"
Name - take this from Brief citation from Paper model NB There are "" that have to be escaped with backslash \ otherwise the .ace file is not reading in fine. Brief_citation name coming from new module at /home/postgres/work/citace_upload/papers/get_brief_citation.pm -- J
URL_constructor "<Article_URL>" from Mappings.txt file on tazendra
Database : "<Full Journal Name>" from Mappings.txt file on tazendra
Name "<Full Journal Name>" from Mappings.txt file on tazendra
URL_constructor "<Journal_URL>" from Mappings.txt file on tazendra
Database : "<Publisher_name>" in Mappings.txt file on tazendra
Name "<Publisher_name>" in Mappings.txt file on tazendra
URL_constructor "<Publisher_URL>" from Mappings.txt file on tazendra
.ace dumper at mangolassi at /home/acedb/draciti/oa_picture_ace_dumper/ (actually at /home/postgres/work/citace_upload/picture/ and symlinked here)
called dump_picture_ace.pl
generates pictures.ace and pictures.err (errorfile, always look at this even if it's usually empty) -- J
How the dumper works
Any error will be written in pictures.err in /home/acedb/draciti/oa_picture_ace_dumper/ Daniela always check it. Any output is going to be in pictures.ace in /home/acedb/draciti/oa_picture_ace_dumper/
An error will be written if there are 2 ore more pgid for the same Paper+source.
the first thing the dumper does is to read into the Picture_source file /home/acedb/draciti/picture_source/picture_source and puts that in a hash %urlacc
If the file is ever not there, the script won't work.
my %urlacc; &readUrlacc(); sub readUrlacc {
my $infile = '/home/acedb/draciti/picture_source/picture_source';
open (IN, "<$infile") or die "Cannot open $infile : $!";
while (my $line = <IN>) {
my ($paper, $filename, $urlaccession) = split/\t/, $line;
if ($urlaccession) { $urlacc{$paper} = $urlaccession; }
} # while (my $line = <IN>)
close (IN) or die "Cannot close $infile : $!";
} # sub readUrlacc
if it finds an association with the WBPaperID and the URL_accession it prints the URL accession then it reads the Mappings.txt file on mangolassi (later on Tazendra) /home/acedb/draciti/oa_picture_ace_dumper/ This file contains the mappings publisher the script will skip the 1st line cause is the header. For each of every other line:
splits into tabs to get each field.
1 $pubname (1st column)
2 $puburl (2nd column)
3 $journame (3rd column)
4 $jourfull (4th column)
5 $joururl (5th column)
6 $arturl (6th column)
7 $template (7th column)
If there is no journal name it skips the line. Each of the values in the Mappings.txt file is associated to the Journal name (3rd column). Additionally, for the Database field, we need a Full Journal Name without spaces associated to the Journal Name otherwise will not read into acedb.
For those 7 values if any of them is missing it will give an error line.
It creates
$entry = "Database : \"$stripped_pubname\"\n"; $entry .= "Name\t\"$pubname\"\n"; $entry .= "URL_constructor\t\"$puburl\"\n"; $entry .= "\n";
Juancarlos, I would like to escape any "/" with a "\" for the following columns of the mappings.txt file. 2nd column: $puburl, 5th column: $joururl, 6th column: $arturl. I have already modified the Mappings.txt file on Mangolassi. J Done
which will be displayed only once at the beginning of the .ace file.
e.g.: Database : "PLoS"
Name "PLoS"
URL_constructor "http:\/\/www.plos.org\/"
$entry = "Database : \"$stripped_jourfull\"\n"; $entry .= "Name\t\"$jourfull\"\n"; $entry .= "URL\t\"$joururl\"\n"; $entry .= "URL_constructor\t\"$arturl\"\n"; $entry .= "\n";
also displayed only once at the beginning of the .ace file.
Database : "PLoSBiology"
Name "PLoS Biology"
URL "http:\/\/www.plosbiology.org\/"
URL_constructor "http://www.plosbiology.org/article/info:doi%2F10.1371%2Fjournal.pbio.%S" //added 02-10-2011 Daniela&Juancarlos
This is it for reading the Mappings.txt file
list of postgres tables: anat_term goid nodump persontext urlaccession chris description lifestage paper remark croppedfrom exprpattern name person source contact
each of the OA fields in the list maps to a pic_ table (see mapping of OA fields to postgres tables chapter in wiki for mappings)
At this point we read the data from postgres as long as there are data in the table for each entry.
After reading into the data it replaces all the new line (line breaks) with a space (e.g. in the remark field if I enter text in separate lines it will display it in the .ace file on the same line)
Then it is creating a mapping from PersonIDs to standard names.
For each paper we are also getting a mapping of the PaperID to
journal
year
title
the first author (if there is also a second author it will add et al.)
starting to dump
The dumper looks at all entries that have a PictureID and for each of them it does the following:
get its pgid. If that pgid has a NO DUMP value it will skip it.
then it will create 2 objects (at the same time):
- Picture object
- Database object
Then the dumper will:
$entry .= "Picture : \"$data{name}{$pgid}\"\n";
if ($data{description}{$pgid}) { $entry .= "Description\t\"$data{description}{$pgid}\"\n"; }
if ($data{source}{$pgid}) { $entry .= "Name\t\"$data{source}{$pgid}\"\n"; }
if ($data{croppedfrom}{$pgid}) { $entry .= "Cropped_from\t\"$data{croppedfrom}{$pgid}\"\n"; }
if ($data{remark}{$pgid}) { $entry .= "Remark\t\"$data{remark}{$pgid}\"\n"; }
meaning if there is a picture object will create a picture object header
e.g. Picture : "WBPicture0000000004"
Juancarlos could you please add the following rule? if pgid and WBPicture number don't match numerically should give an error. Thanks D
when dumping an object you'll get an error if the name has spaces in the front or at the end (very important, because other objects linked to this will also have that space, so you should remove any connections to that object ID, fix the spaces, then remake the connections to the fixed object ID), and/or if the number of the object id isn't the same as the pgid -- J
Juancarlos could you please add the following rule? What is in the "Source" field should be unique. There should never be 2 objects with the same source. D Done, there's already one error of that type, and 2 errors of the pgid not matching object ID type -- J Great thanks! Work fine D
if there is description it will create a Description .ace tag
e.g.: Description "Confocal images of mid-stage embryos expressing GFP under the control of the wild-type K07C11.4 promoter. Percentages are the fraction of transgenic embryos expressing GFP; the remainder of embryos do not express GFP. Dashed lines indicate the outline of the developing pharynx."
Added the following rule: if there is more than one space it will be converted in a single space. Line breaks are deleted. 02.09.2011
"" symbols are escaped: from J: I'm not escaping it on everything, because the multiontology fields use "," to separate different values, so it's easier to not mess with it. Given that they're controlled vocabulary, I've made it filter on description, source, remark, and urlaccession (not persontext, but we could add it there, it was just additionally extra code more than the rest, and I wasn't sure it was necessary).
same for source, cropped_from and remark.
if ($data{exprpattern}{$pgid}) {
my ($data) = $data{exprpattern}{$pgid} =~ m/^\"(.*)\"$/;
my (@data) = split/\",\"/, $data;
foreach my $data (@data) { $entry .= "Expr_pattern\t\"$data\"\n"; } }
if ($data{goid}{$pgid}) {
my ($data) = $data{goid}{$pgid} =~ m/^\"(.*)\"$/;
my (@data) = split/\",\"/, $data;
foreach my $data (@data) { $entry .= "Cellular_component\t\"$data\"\n"; } }
if ($data{anat_term}{$pgid}) {
my ($data) = $data{anat_term}{$pgid} =~ m/^\"(.*)\"$/;
my (@data) = split/\",\"/, $data;
foreach my $data (@data) { $entry .= "Anatomy\t\"$data\"\n"; } }
it does the same as above with the difference that every anatomy entry (or GO entry, or Expr_pattern) will be displayed in a separate line: e.g:
Anatomy "WBbt:0003681"
Anatomy "WBbt:0005175"
It looks at the contact info, it dumps the Contact e.g. Contact "WBPerson12345"
If more than one contact does the same as Anatomy (1 entry per line).
If there is a contact -> Then it generates a template text: "WormBase thanks <Person_name> for providing the pictures." NB: remember that the dumper will use the Person_name to generate the acknowledgement, not the Contact field. Whenever pictures are submitted from persons other then publications the contact field and the person field should both be filled. (DR 110324)
Now the dumper looks into Person data
if there is a Person and Person Text -> it will join all <pic_person> objects's starndard names with commas, then comma, then <pic_persontext> text. During the curation: If the Person field is filled and you need to add something in the Person Text you should do the following: If it is a single entity in the text write e.g. "and Paul Sternberg's lab" if there are multiple entities e.g. summer student 1 (non WBPerson), summer student 2 (non WBPerson), and summer student 3 (non WBPerson) write "John Smith, George Brown, and Mike Lee".
otherwise if there is person data will
For one person: Juancarlos for 2 people: Daniela and Juancarlos For 3 or more people: Daniela, Juancarlos, and Jim
This is the same mapping as before when it converts WBPersonID into standard name
Otherwise if Person_text writes only the person text. Free text. For one person: John Smith. For 2 persons: John Smith and Mark Brown. And so on.
If there is a Reference (pic_paper table in postgres) it dumps the Reference:
e.g.: Reference "WBPaper00024505"
then if there is a year it will generate a Publication_year:
e.g. Publication_year "2004"
If it does not have a publication year it will generate an error (Daniela will fix it according to Kimberly's rules) and it will say BLANK in the brief citation
If there is not a Journal it will give an error
If there is a Journal:
if there is not a mapping to the mapping file it will give an error
if there is a Journal and there is a mapping and there is Full journal with stripped spaces it will print:
it will print "Journal_URL\t\"$mappings{strippedjourfull}{$journal}\""
e.g.: Journal_URL "PLoSBiology"
if there isn't a stripped Journal name it will give an error
It will do the same thing for the Publisher_URL
it will print: "Publisher_URL\t\"$mappings{strippedpubname}{$journal}\"\n"
Publisher_URL "PLoS" Daniela add stripped name as for Jounal_URL
if there is a Journal and there is a mapping and there is a URL_accession it will print:
if ($data{urlaccession}{$pgid}) { # new output line for Daniela 2011 02 22
my ($urlaccession) = &filterAce($data{urlaccession}{$pgid});
$entry .= "Article_URL\t\"$mappings{strippedjourfull}{$journal}\" \"id\" \"$urlaccession\"\n"; }
elsif ($urlacc{$wbpaper}) {
my ($urlaccession) = &filterAce($urlacc{$wbpaper});
$entry .= "Article_URL\t\"$mappings{strippedjourfull}{$journal}\" \"id\" \"$urlaccession\"\n"; }
else { print ERR "$pgid no urlaccession for $wbpaper\n"; }
e.g.: Article_URL "PLoSBiology" "id" "0030053"
home/acedb/draciti/oa_picture_ace_dumper/dump_picture_ace.pl now reads /home/acedb/draciti/picture_source/picture_source If the file isn't there or can't be read there's an error message and the program stops. Data from the 3rd column (always the third column) gets associated with data from the 1st column. If there's no data for the pg table pic_urlaccession, it looks to see if there's a match from the pic_reference table to the mapping on picture source, and uses the data from the third column as the urlaccession. Otherwise it gives an error.
If there isn't take the Accession number from the picture_source file (where there is mapping paper <-> Acc No)
if there is no Accession number in any of the 2 will give error
if there is a Journal and there is a mapping and there is a template it will print the template:
Template "WormBase thanks the journal <Journal_URL> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Journal_URL>, <Article_URL>. Copyright <Publication_year> with permission from <Publisher_URL>"
if not will give an error.
The following part of the dumper has been deleted after Paul Davis suggestions (feb 9th 2011) as the URL for the paper has been added to the Journal Database and the Brief citation will be pulled out directly from the ?Paper object by the webteam.
if there is a Journal and there is a mapping and there is a URL_accession it will print:
Article_URL\t${wbpaper}_URL id $urlaccession
e.g.:
Article_URL WBPaper00024505_URL id 0020352
it will also creat the database object for the Article_URL
it will be
"Database : ${wbpaper}_URL"
If there is an Article_URL it will print
URL_constructor\t\"$mappings{arturl}{$journal}\"
my ($brief_citation) = &getBriefCitation( $firstauthor, $year, $journal, $title ); # from package /home/postgres/work/citace_upload/papers/get_brief_citation.pm Name\t\"$brief_citation\"
There are "" that have to be escaped with backslash \ otherwise the .ace file is not reading in fine
e.g.: Database : WBPaper00024505_URL Name "Gaudet J et al. (2004) PLoS Biol \"Whole-genome analysis of temporal gene expression during foregut ....\"" URL_constructor "http:\/\/www.plosbiology.org\/article\/info:doi%2F10.1371%2Fjournal.pbio.%S"
Symbols conversion in the dumper script
While dumping the following symbols will be converted:
µ in u
± in +- change it in +/-
" are escaped in the following fields: description, source, remark, and urlaccession
multiple spaces in the "Description" section will be converted in to one single space
μ converts in u
α converts in alfa change it in alpha
¡C converts in C
° converts in C
Ð converts in -
′ converts in '
¼ converts in u
± converts in alfa change it in alpha
âÂÂ1 converts in -
> converts in >
< converts in <
â² converts in '
β converts in Beta
âÂÂ¥ converts in ≥
∼ converts in ~
âü converts in -
² converts in Beta (e.g. pgid 7493)
Ï€ converts in ∏
⬠converts in ∏
± converts in alpha
¼ converts in u
â converts in Delta
â⬲ converts in '
ââ°¥ converts in ≥
Ãâ converts in x
0Ãâ converts in x
° converts in °
â˼ converts in -
â⬲ converts in '
° converts in °
â⬲ converts in '
¼ converts in u I think we put this already but dumps it wrong ((e.g. pgid 7528 7659 and several more)
Web display
Adrian's code has already accounted for the contact. If there is both a paper reference and a contact, both will be displayed on the paper page. When there is no reference and there IS a contact, then on the expression pattern page, the contact will be acknowledged by "Courtesy of <Person>". A
Notes
J: I forgot to point that when doing the WBPicture object creation, if you need to edit the name of a picture object, you should be careful not to have extra spaces around it or extra / missing digits, and that the uppercase / lowercase is all okay. If you associated something as being cropped to WBPicture0000000001 (or later under Expr_pattern), and you then want to change it to WBPicture0000000002 don't forget that you changing the object ID will _not_ change all associations to it, so if picture 5 is still associated with picture 1, you'll still have to query picture 5 to change its associated picture 1 to a picture 2. and also it's an ontology, so if picture 1 is no longer a valid picture, it might not come up in the OA to change, so you'd have to delete all associations to picture 1 first. then change 1 to 2. then reassign those associations to picture 2.
I imagine this will almost never be a problem for picture objects (like it is for genes being merged and split), but you should be aware of it. (and probably ask about if it's not clear, and put it on the wiki in some section about renaming objects or something like that). If you change the name right after duplicate, you wouldn't have associated anything to it, so it would be okay. And I imagine that's what you'll mostly be doing, so it should be okay.
Draft OA for picture curation
WBPicture "" // this will be the picture ID -> generates automatically upon entry. We should have a "duplicate" button which generates a new ID. The object ID for the name reflects the postgres ID (pgid). Actually, the way the code is laid out, duplicate cannot assign a new pictureID, it has to duplicate the existing object ID. OK, no problem, I will do as Karen does! I was thinking of the way that date_last_updated changes in the GO config, but even then for duplicates it duplicates the old date, sorry =( But if the picture ID will always be the postgres ID, you can change the number based on the number in the pgid field. When Karen creates a new molecule object (only other config that creates IDs automatically), she still has to change the name too. What should the IDs look like ? It should be WBPicture0000000001 and progressive numbers --D Ok, changed from WBPicture:12345678 to WBPicture1234567890 (no : and 10 digits) Daniela has double checked with Gary -> Is OK to remove colon, which is used mainly for ontologies -- J J can you please change the Name into WBPicture in the OA? Done - J
Reference "" // This will record which paper this picture comes from -> ontology This is getting expr_pattern data from table obo_data_pic_exprpattern TODO change this when Expr Pattern OA is live. Make note on wiki for Expr Pattern OA -- J. OK J when I start the Expr_pattern OA wiki I'll make a note TODO Daniela--D Get the Jpgs from the picture_source file in Tazendra Daniela specify path for J. For now I want to be part of the automatic term info update as opposed to manual. I put the file "picture_source" on tazendra under draciti J please add Journal_name and Publication_year. And always show those 2 fields so that it is clear for me when is missing D TODO on tazendra create obo_ tables for pic_picturesource at /home/postgres/work/pgpopulation/obo_oa_ontologies/create_obo_pic_picturesource.pl -- J done 2010 12 21 Have moved picture_source file on mangolassi to /home/acedb/draciti/picture_source TODO on tazendra, incorporate to cronjob /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl -- J done 2010 12 21 Reference Term info now always has picture_source .jpg files listed, as well as Journal and Year (or BLANK for journal / year if not available) Rewrite /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl to use new format generated by mergeToOICR.pl script on Daniela's computer after Daniela makes it clear how to deal with .txt or .docx or what-not. -- J Juancarlos I have converted all the .docx files into .txt files. I would like to see a link to the .txt file in the term info (same place where I see the file names). If I understood correctly the mergeToOICR script should be changed including the .txt and then I will do the same as before. Scp to Tazendra and rsync with Canopus I've updated the script on canopus, so that it should work if you copy it to your computer (replace the one there, but maybe keep a copy just as a backup), and copied the new picture_source to mangolassi and populated the term info. But there are no WBPerson entries, so I can't test if that's going to work in the Term Info for contact. -- J I Copied the script on my computer, no problem that we cannot test the WBPerson Term Info yet. everything seems to work fine D We should add the locus information displayed together with the Expr_pattern in the Paper term info display. To do that: For every expr_pattern, look at the expr_pattern ontology to find Gene Name then use same mapping as GO OA to map gene to locus. If there is no locus put synonym. Thanks :) paper term info's expr now maps to a wbgene from obo_data_pic_exprpattern, and that maps to a locus from gin_synonyms gin_seqname gin_locus J
Update from Nov 23rd. D and J agreed that the pictures will be put on Canopus and J will take the picture source from there: /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl adapt this script to once a day crawl all the picture folders on canopus to get mapping of WBPapers to all their picture. Need to know what the picture folders are, and they need to be viewable from a webbrowser.
Contact "" // person multiontology on people as in Phenotype TODO update person term info to have contact from obo_data_pic_picturesource once Daniela tells me how to parse the file for .txt / .docx -- J. Same as for Reference D No data in Canopus with WBPerson, so can't test to see if it works -- J OK D Displaying term info in Contact field Fix script to populate the obo_data so that it will display file names in term info (.jpg and .txt)D
Description "" // this will be figure legend -> big text
Source "" // this will be the actual picture name -> small text J, when you are in the office, I will show you a file I might use for autocomplete the source. i don't know if it is feasible but we should have a look at it cause it can save me lots of copy-pasting ^^ If it is not ok we keep the small text field D okay, but if you want a set of stuff to autocomplete, you'd have to maintain something for the database to update from it. for example, there's a lot of obo files maintained in cvs in sourceforge, so there's a script that updates based on those. if you wanted to commit your file to some cvs repository like that, it should work -- J. OK once you see the file in the office you can tell me how easy it is to maintain that file -- D Ok, sure. It's more an issue of where you're going to keep it for a script to pick it up - J Daniela todo -> scp on Tazendra a file the file Picture_source and update it constantly every time you are done with a journal J I scp in tazendra under draciti a file called picture_source. D Oh, sorry, when we're live it should be on tazendra, for now since we're working on stuff I'm putting it on mangolassi. So you wanted to run a script manually to populate this (extra step) or did you want a cronjob to pick it up and update everyday (potential for 24 hour delay before you can curate, when would you want it to run ?). To be clear, what should I do with this file ? J If I would like to have a cronjob (update every day). If I understand correctly I will modify the file once in a while (whenever I have new data to put in) and the cronjob will automatically update the tables. If that is the case, let's go for it! Automatic, yes, but not instantaneous, I just want to be clear that if the script runs at 2am every day, and you update it at noon, you won't get to see the term info updates until the next day at 2am -- J No problem D. For Reference field, only display the JPGs when entering a paper. J you mean displaying only the JPGs coming from the picture_source file and not displaying the .docx files right? if this is what you mean the answer is yes. And if I am correct, in the Reference field, I will still continue to see expression pattern data, correct? D Yes, sorry, I meant just the filename of the jpg files, as opposed to the other files, and yes, in addition to other WBPaper data -- J OK D For source it doesn't matter because it's text, not ontology ? yes D For cropped_from, ignore it because it will autocomplete from this OA Source field, not from the picture_source information we get from this file ? Is all that correct ? -- J I think so.. Let's talk about this after the meeting to make sure, then you can confirm on this wiki -- J OK D now that I am a bit more free from the modelling I can finally seriously testing the OA. And regarding this, this morning is not working, maybe because you are working on it? ^^ The error I get is JSON parse failed D Sorry about that ! I had to wipe and repopulate the database on the sandbox for some interaction stuff a few times, and I forgot to recreate the picture tables afterwards (also, sorry, any data you entered is gone) - J No problem D
Cropped_from "" this will only be used by the cropped images to indicate its mother picture -> ontology of picture objects Single ontology. Have not done this yet since there are no real picture objects yet. What do you want to show in term info here? The cropped from will be used only for duplicated objects. Let's say I have a mother picture and I want to duplicate it because I have a cropped panel. I would like to see Cropped_from "journal.pbio.0020352.g006" You actually want the WBPicture ID here, right ? -- J. I want to have the same name as "source" of the mother pictureD What should it autocomplete on? It should autocomplete on the "source" of the mother picture --D Do you want anything on the term info, just the name and ID ? autocomplete only on source, not both source and ID ? If you can autocomplete on both source and ID would be good!D Sure (do reply to the PictureID stored in postgres, I'm pretty sure that's what you want, but do confirm. OK Juancarlos, maybe I confused myself. To summarize the Cropped_from field: In the Cropped_from field I want to autocomplete with the "source" of the mother picture. In the Term info I would like to see The name and the ID (e.g. WBPicture0000000001, and journal.pbio.0020352.g006). Does that sound right to you? otherwise I'll show you on Thursday -- D It kind of makes sense, but I'm not sure it's good. Each picture object has a picture ID, so when referring to it, it's best to refer to the picture ID, because it's potentially possible that the name could change. Otherwise we'd just have picture names instead of picture IDs, right ? So I think of the source as a name, and we could have this field autocomplete on the source/name, but then store/save the picture ID. Then when dumping to .ace outputting the source of that picture object. This way if you make ID 1 -> source "blah", then ID 2 -> cropped from ID 1, then change ID 1 -> source "different", when you dump picture 2 it would say cropped from picture ID 1, with source "different". If in the same case you put in picture 2 source -> "blah", when you dumped picture 2 it would always say "blah". Does that make sense ? -- J In the Cropped_from we will autocomplete on Source (file name) then WBPicture ID; and store in postgres the picture ID -- D Right, and we'll do that based on the Source OA field / postgres table, not what was entered in the "picture_source" file above -- J Yes! D Autocomplete on source then WBPicture name/ID. Show on Term Info name/ID, source, reference. -- J
Expr_pattern "" // this relates to the Expr_pattern associated with the picture -> multiontology File that has Expr_pattern <-> paper association is ExprWS221.ace received from Wen October, 22 2010. In term info we'd like to see Gene, Pattern, Reference, Reporter_gene, Life_stage Anatomy_term, GO_term. Autocomplete just on Expr_pattern ID. For Anatomy_term retrieve Anatomy_term ID and name from app_anatomy. Created obo_<data|name>_pic_exprpattern tables temporarily until Expr_pattern OA is live. TODO get rid of these tables once Expr_pattern OA is live. See below Make note on wiki for Expr Pattern OA. Created in mangolassi at /home/postgres/work/pgpopulation/exp_exprpattern/ When live on tazendra, TODO use /home/postgres/work/pgpopulation/exp_exprpattern/create_obo_pic_exprpattern.pl and populate_obo_exprpattern.pl DONE created on tazendra for grg_generegulation, which needed these tables. Also, since you requested the Expr_pattern field to be an ontology field, you should make sure that it works for you and Xiaodong's data, then tell her that if she wants it to work like that in her OA, she needs to let me know and then we need to transfer her data from text to ontology or multiontology. OK< Xiaodong said it is fine with her --D
Expr_pattern term info
Juancarlos, here is what I want to see in the picture OA for Expression Pattern term info
- Expr id : exp_name -- autocomplete on name only
- Gene : exp_gene -- show WBGenename, locus and synonym as in expr Pattern OA. e.g.: Oh, but this is a multiontology. Do you want to see all this info for all these genes in separate blocks ? I don't mind how is displayed actually, if it is easier for you could also be: whichever you like, I just thought it was confusing the other way, if everything is one line per data, then it's more clear in that sense
"WBGene00022781, pmt-1, ZK622.3, phi-40" "WBGene00017066, maco-1, D2092.5"
It will be clear from the context that synonym means the synonym for that ID only ? I imagine every gene that has a synonym should have a unique synonym that'd be nice, but some genes are synonyms of other loci. You can tell from the paragraph context, but if we're doing it one line per wbgene, it's probably clearer
id : WBGene00022781 locus : pmt-1 synonym : ZK622.3 synonym : phi-40
- Anatomy_term : exp_anatomy exp_qualifier exp_qualifiertext --display all data one data per line e.g.: so for exp_anatomy show the name and the ID in doublequotes, yes and the other two things repeated for each term in exp_anatomy. the repeated things were exp_qualifier and exp_qualifiertext. I changed the example so maybe is more clear.
Anatomy_term : "nerve ring is WBbt:0006749" Certain expressed in XYZ Anatomy_term : "pharynx is WBbt:0003681" Partial expression detected in a subset of neurons
- GO_term : exp_goid -- show GO ID followed by the GO name (I checked in GO OA and what I want to see here is "name" , I don't know how it is called in the GO tables e.g. ok, I'll figure it out great
GO_term : "GO:0005737" Cytoplasm
- Subcellular Localization : exp_subcellloc
- Life_stage : exp_lifestage -- Convert the life stage IDs into names from the obo_name_lifestage e.g. "L3 larva" so name only, no ID correct
- Antibody_text : exp_antibodytext
- Reporter Gene : exp_reportergene
- In_Situ : exp_insitu
- RT_PCR : exp_rtpcr
- Northern : exp_northern
- Western : exp_western
- Antibody_info : exp_antibody so this is the multiontology field, and the data looks like : "[cgc3991]:apr-1_b","[cgc3991]:apr-1_a" is good to have them in one line
- Pattern : exp_pattern
- Transgene : exp_transgene data looks like "kyIs136","kyIs131","kyIs137","kyIs140" fine
- Reference : exp_paper data looks like "WBPaper00001926","WBPaper00001469" fine
No western ? good catch!! I missed it :p np =)
Remark "" // For other curator notes -> big text Remark should be dumped as text. Try to generate few data in the OA and to dump a .ace file. See what will be dumped as remark. Tried (nov 28 2010) it works fine.
Cellular_component "" // For sub-cellular localization -> multiontology of GO_Term like gop_goid. File that has Expr_pattern <-> GO_term association is ExprWS221.ace received from Wen October, 22 2010. After I copy wen's file on tazendra and tell you to parse it MAKE PAPERS TERM INFO DISPLAY EXPR_PATTERN
Anatomy_term "" // It will link the picture object directly to an Anatomy Object -> multiontology. Should work like app_anatomy File that has Expr_pattern <-> Anatomy_term association is ExprWS221.ace received from Wen October, 22 2010. File that has Anatomy_term <-> anatomy name association is http://obo.cvs.sourceforge.net/viewvc/obo/obo/ontology/anatomy/gross_anatomy/animal_gross_anatomy/worm/worm_anatomy/WBbt.obo For example, Wen's file has Anatomy_term "WBbt:0004854" and OBO file has WBbt:0004854 name: vm1 def: "Vulval muscle 1"
Article_URL_Accession "" // Text this is the unique id pointing to a paper URL
Person "" person multiontology on people as in Phenotype
Person_text "" free text small. Note on the .ace file for Person and Person_name: join all person objects's starndard names with commas, then comma, then person_name text. If there's no person_name text. join all person objects's standard names with commas, except for the last one, which is joined by "<comma> and ".
Life_stage "" multiontology autocomplete on ExprWS221.ace received from Wen October, 22 2010 (same file as others) --D I don't understand what you mean about the Expr file, but you can see a life stage field in the phenotype OA, and see if that works like you'd want -J Yes, it si perfect to have the life stage field like in the phenotype OA D Ok, We have a few new tables to make, so I'll wait until we're set with those and make them all at once (curation_status, lifestage, anything else ?) -- J Give me a couple of days to test it more and to solve the Acknowledgment issue. I did not hear from the webteam yet and I feel bad coming back and forth to you with new requests! :( That's okay, we can wait for the acknowledgements, but are those the only things you want for non-acknowledgements ? Just curation_status and lifestage ? No. Have created tables urlaccession person persontext lifestage nodump chris 2010 11 15 Please test OA -- J Tested D
Curator "" field, where do you want it ? -- J Where you put it it is totally fine :) -D
No dump "" Toggle J please can you add the no dump field? Thanks D
Chris "" Toggle
Acknowledgments ""// the acknowledgment field will have more than one tag
* Template Text Daniela created a table with Publisher/template text association -> Mappings.txt tab delimited file
* Publication_year J will get data from paper tables
* Article_URL Daniela created a table with Journal name/URL constructor -> Mappings.txt tab delimited file
* Journal_URL Daniela created a table with Journal name/Journal_URL -> Mappings.txt tab delimited file
* Publisher_URL Daniela created a table with Publisher/Publisher URL -> Mappings.txt tab delimited file
* Person_name will have 2 boxes in OA
where
"Template" // is the template sentence e.g. "WormBase wishes to thank the journal <Journal_name> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Journal_name>, <Article_URL>. Copyright (<Publication_year>) with permission from <Publisher_URL>." The template sentence will change accordingly to what publishers need but the tags populating it will always be the ones listed below Okay, it sounds like you don't want to store the template in the OA, you'll just tell me to hardcode in the dumper script that if it's a given Journal, use this template, if another some specific other template, and so forth ; so that if the template ever changes for a given journal we can just change the dumper script, correct ? - J Supercorrect! -D Also, you should probably look at the full list of Journal objects, because there are _tons_ (1404) due to minor differences in spelling and what-not. You probably can't map all those to a template, but I don't know. Maybe once we have a list of papers (do we already ?) we can see what journals exist for those given papers. J I have a list of 184 journals containing Expr_pattern data but I don't have yet a number for the template sentences because I am still working on getting copyright and permissions. It can be there will be 5 or 50, I just don't know yet - J Right on, got it -- J You also need to work out the issue of pubmed_final with Kimberly (see Journal_name section) Here's the postgres query for the 1404 journals SELECT DISTINCT(pap_journal) FROM pap_journal ; which you can see on the referenceform.cgi linked in the sitemap -- J OK I wrote to Kimberly, her answer is that if a paper has a PMID and is missing Journal or Year, let her know. If it doesn't have a PMID and is missing that information, I should feel free to fill it in using the paper editor.
"Journal_name" // we can retrieve it from the ?Paper data model (?Paper Reference Journal UNIQUE ?Text) Yes, but you need to tell me what to do if there's no Journal info for a given paper. I need to write something in the code of the dumper script to account for cases where there's no Journal. I see. If you're entering a paper you can see that there is or isn't a journal, but if the pubmed_final field is not set to final, then seeing a journal doesn't always mean that there will be a journal later. You should talk to Kimberly about this. - J I wrote to Kimberly and asked her how we should proceed on that. I'll let you know as soon as she gets back to me D Hopefully we can talk to her after the conference call tomorrow -- J see Kimberly answer above D J please get data from Paper tables and If the journal-name is empty write BLANK D
"Publication_year" we can retrieve it from the ?Paper data model (?Paper Reference Publication_date UNIQUE ?Text) Same as above -- J Same as above -- D J please get data from Paper tables and If the Publication_year is empty write BLANK D
"Article_URL" // this will contain the URL pointing to the paper citation. Do you store that in postgres, or can we generate this from a paper ID pointing to WormBase ? -- Daniela will create tables journal name -> Article_URL
"Journal_URL" // this will contain the URL pointing to the journal. Do you store that in postgres, or can we generate this from a paper ID pointing to WormBase ? -- Daniela will create tables journal name -> Journal_URL
"Publisher_URL" this will contain the URL pointing to the publisher's homepage. Are the mappings always the same that we can get based on the journal name ? -- J Daniela will generate tables Publisher -> publisher URL
To go live on tazendra
psql -e testdb < /home/postgres/work/pgpopulation/pic_picture/create_tables done 2010 12 21
/home/postgres/work/pgpopulation/obo_oa_ontologies/create_obo_pic_picturesource.pl done 2010 12 21
/home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl done 2010 12 21
/home/postgres/work/pgpopulation/pic_picture/parse_pictures.pl > logfile done 2010 12 21
set to cronjob (everyday at 2am) 0 2 * * * /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl done 2010 12 21
tazendra picture OA is now live, verify that everything works properly then set the issue as resolved 2010 12 21
(probably manually edit some entries that weren't parsed properly)
Bugs and Fixes
J added Chronograms names in the Expression pattern field in OA for old picture objects -> March 22nd 2011
Pipeline for picture handling Development
- set up the picture files handling as we discussed on skype. I would like to have the files on my computer and rsync it to canopus. On my computer I have a folder named Canopus/Pictures which contains 3 directories: 200/, 600/ and OICR.(I put the same thing on canopus for you to see /Users/danielaraciti/Desktop/Canopus/Pictures) I would like that all files in the 600/ will be renamed having a _600 extension before the .jpg. E.g.in directory called WBPaper00024399 the file called journal.pbio.0020280.g007_A.jpg should be named journal.pbio.0020280.g007_A_600.jpg.
Then I would need to have the files from /600 and /200 and put them all together in a new folder called OICR (run script ./mergeToOICR.pl). The script takes files from 600/ and 200/ and moves them in Pictures/OICR. Once you run it it generates a file that you should scp into tazendra /home/acedb/draciti/picture_source/picture_source
mergeToOICR.pl script takes from Pictures/ there is 200/ 600/ OICR/ From 200/ take all folders and move to OICR/ From 600/ loop through each directory, and for each WBPaper####/ rename all pictures replacing .jpg with _600.jpg and move to appropriate OICR/WBPaper####/ directory. Do a check that all pictures in OICR/WBPaper####/ have a normal, _200, and _600 version. Also writes file picture_source with mappings of WBPaper to source.jpg names for the script that populates the appropriate .obo tables. Daniela will scp it to the proper location (/home/acedb/draciti/) after running the mergeToOICR.pl -- J
Script is in: /Users/danielaraciti/Desktop/Canopus/Pictures/mergeToOICR.pl
Then we rsync Pictures/OICR/ to Canopus.
rsync --progress --delete-after -a /Users/danielaraciti/Desktop/Canopus/Pictures/OICR/ daniela@canopus.caltech.edu:OICR/
Once this is done the 200/ directory should be empty and the 600/ directory has a list of empty folders which should be discarded. The next time I have a batch of pictures I recreate 600/ and 200/ and rerun ./mergeToOICR.pl
The very last step of the process is to adapt the following script as you described and to add in term info the .txt files. J did it on Dec 6th We now have a link in Reference and Contact to the .txt files.
/home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl adapt this script to once a day crawl all the picture folders on canopus to get mapping of WBPapers to all their picture. Need to know what the picture folders are, and they need to be viewable from a webbrowser. Done
Reading in existing picture objects
mangolassi : /home/postgres/work/pgpopulation/pic_picture/parse_pictures.pl
J: To make sure it works, we need to read in the existing picture objects from acedb, so scp the source file with the .ace objects you want populated to tazendra, and write somewhere on the wiki, how I should map the .ace data to these OA tables.
D: The file with existing picture objects is in tazendra and is called citace220picture.ace
in the file you have only 2 info for a single object:
Picture : "29055F14H3.11_1.jpg" -> this should map to the "source" field in the OA
Expr_pattern "Expr7505" -> this should map to the "Expr_Pattern" field in the OA
we need to add
Picture ID for each of them WBPicture0000000001, WBPicture0000000002 and so on
Reference "WBPaper12345678"
Hm, I'm not following. The data only has those two thing (jpg and expr_pattern), so where does the other data come from ? /
Sure sorry, I thought you could take it from the expr_pattern data. I put another .ace file on tazendra called ExprWS221.ace where there are the Expr_patterns <-> Paper IDs. If this does not make sense at all, just let me know :) What about the Expr_pattern objects that have multiple papers ? -- J There should be no case in which an Expr_pattern object has multiple papers associated D At a glance, Expr_pattern : "Expr12" has Reference "WBPaper00001926" Reference "WBPaper00001469" I haven't programatically looked into it though -- J J Please use WBPaper00001926 as reference for Expr12. done - J Comment: We are using the latest reference whenever there are 2 references associated to a picture. There were only 2 cases: Expr12 and the large scale study mentioned below : there is double correspondence for 2 papers, WBPaper00006525 and WBPaper00031006, for one single expression pattern and this is happening because there are 2 large scale studies but is anyway kind of weird. I spoke with Wen (who curated the data) and Raymond and we came up with the following consensus: we should associate to all those objects the reference WBPaper00031006 because is the most recent paper. If the second paper is WBPaper00031006, that's now the only paper, seems okay -- J OK --D
Look at mangolassi : /home/postgres/work/pgpopulation/pic_picture/errors There are many Chronograms which have no mappings to WBPapers, and many Expr_pattern objects that map to two papers (the same pair for all of them) -- J J thanks for looking into this. I checked Acedb and talked to Wen, all the chronograms are coming from a large scale study: Dupuy D et al. (2007) Nat Biotechnol "Genome-scale analysis of in vivo spatiotemporal promoter activity in ...." WBPaper00029359 I dumped a .ace file for chronograms, if you want to have a look at the file is on Tazendra in draciti and is called Chronograms.ace. I'm now also comparing against that file Great, thanks!
After this keep a hold on existing picture objects as there are pictures from Flickr and there are still some issues I have to resolve with Todd and Raymond. Thanks! D What do you mean by keeping a hold on existing picture object ? I don't understand -- J I mean that we should not dump any .ace file and put it in acedb yet. We want to generate new data and see if the process works fine with the new model --D I won't be dumping any data, I'll just make the dumper, and it's up to you to put it in acedb when you want to, but keep in mind that all data that gets populated in postgres will be dumper unless it has the NO DUMP flag. -- J OK, I just wanted to make sure. I think we discussed already that all the existing picture objects should be flagged as No dump--D
The file still has errors for Expr without reference. You can see it on mangolassi. Thanks Juancarlos, if there is no reference leave the "reference" field blank. This issue raised another problem, of course... :) J We need a new field in the OA called "Contact" a person multiontology on people as in Phenotype. It should be between "Reference" and "Description" and in the final.ace file should be dumped as Contact "standard name from ?Person" e.g. Contact "Raymond Lee". Please let me know if this is clear. I have put the description in the "Draft OA for picture curation" chapter and in "Final .ace file should be dumped as". I thought you were going to create persons that were personal communications, or some such, after talking to Kimberly. Don't you need a WBPaperID to be part of the Name field and Article_URL ? There is no Author field. -- J Juancarlos, I thought this through, I checked the Paper Data Model and pondered the pros and cons of having one solution over the other. I also asked Raymond an advice on that. The cleanest way to go is to add in the ?Picture model a Contact tag and therefore adding a Contact tag in OA. I hope this does not cause you too many problems. We can talk on Skype if you wish to. I have manually checked the file with the Errors (Expr without reference). Once we have added the "Contact" tag in the OA we will populate it as described below. I know the picture OA procedural way was a bit more challenging because we were developing the model in parallel with the OA but we are really almost there and I am extremely grateful to you for everything! So to summarize what should be done:
- Create a new field in the OA called "Contact" // person multiontology on people as in Phenotype. It should be between "Reference" and "Description" and in the final.ace file should be dumped as Contact "WBPersonID" e.g. Contact "WBPerson12028". Done
- Populate all the Expr without reference. All the entries that gave error should have WBPerson266 (Ian Hope) in the "Contact" field except:
b0523_5_phx.jpeg Expr35, b0523_5_vul.jpeg Expr35 which should have WBPerson1232 (Lynch AS)
c07b52v.jpeg Expr83, c07b54ec.jpeg Expr85, c07b54la.jpeg Expr85 Which now have a reference WBPaper00002319 (at that time it was in press)
J: I thought there was always one Expr per picture, but that is not the case :
- c09f9_3_larv.jpeg has many expr Expr2060 Expr2006
- Expr3072_3073.png has many expr Expr3072 Expr3073
- f59b2_13_can.jpeg has many expr Expr19 Expr8
- f59b2_13_head.jpeg has many expr Expr19 Expr8
- zc84_3_all.jpeg has many expr Expr25 Expr26
- zc84_3_vnc.jpeg has many expr Expr25 Expr26
And Expr19 has a paper, but Expr8 does not. Expr25 has, Expr26 does not.
I have checked this. It can well be that there is more than one expr_ per picture. However I don't know why Expr19 has a paper and Expr8 does not. I can fix this manually when we are live on tazendra. Fixed. Expr 8 is associated with Hope IA (1991) Develoopment 'promoter trapping' in Cenorabditis Elegans. 110329 DR
Also, 2 entries have description, it's probably easier if you enter those descriptions manually later if you want them (when it's live). For that matter, maybe the script can just populate the ones above and you can also fix those manually when it's live, since there's only 6 pictures. I think the best is that I fix everything manually when it's live. Can you just write here the ids of the 2 that have description? D Sounds good. No, the pgids could change if we change things or who-knows-what when we go live on tazendra. Just look at the .ace file, search for that tag, then query those objects when it's live. -- J OK D. The 2 objects that have a descroption associated are Expr_3071jpg and Expr3072_3073.png. Those were deleted from OA-- see below 110329 DR
Wiped the tables and populated this data by calling /home/postgres/work/pgpopulation/pic_picture/parse_pictures.pl
J: So the OA will have a Person field, Person_text field, and Contact field, all of which refer to people? D: Yes. the reason for that is that the dumper constructs a sentence for the acknowledgments taking the Person field and combining it with the Person_text field. Therefore in acedb the Person_name is a text. We need instead something separate from the acknowledgment which refers only to the contact author, in case somebody wants the original file. That is the reason of why having the Contact tag
J: How does the dumper change to deal with ?Picture objects without a Paper ? (and what if they have no paper and no contact) D: If there is no Reference it stays blank. There will be no such case in which we have no paper and no contact
J: Why make contact a multiontology (vs. ontology)
D: Because there could be cases in which the Principal investigators will be 2
J: And why dump the Standard_name instead of the ?Person objects D: You are right we should dump the ?Person objects
J: There's still a lot of stuff that's bold from before.I did not change them back because I thought they are reminders for you. D: the stuff that is bold from before I left it bold because I thought they are comments for yourself, such as: TODO on tazendra create obo_ tables for pic_picturesource at /home/postgres/work/pgpopulation/obo_oa_ontologies/create_obo_pic_picturesource.pl -- J Have moved picture_source file on mangolassi to /home/acedb/draciti/picture_source TODO on tazendra, incorporate to cronjob /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl -- J Reference Term info now always has picture_source .jpg files listed, as well as Journal and Year (or BLANK for journal / year if not available)
J: Please make a separate ticket about the 600 / 200, and put in a different section of the wiki, as it doesn't really have to do with reading in existing picture objects. D: OK
Also, in the future, please copy files to mangolassi instead of tazendra if we're doing the development there -- J Will do :)--D
Do you want me to make up the IDs starting from 1 and going forward ? Yes ok -- J
Where do the Paper IDs come from ? see above
We don't need a year because that's in the paper object, right ? right
We also need a curator, would that be you or someone else ? See below
Any other field that we should fill in that is required ? See below, there was more about the topic it goes down till the To Do chapter:) thanks!
I will add description and anatomy once I'll get to annotate them. You mean you'll enter them through the OA when it's live, not that you'll enter them into acedb, nor into the .ace file, right ? -- J Yes, I will enter them through OA when it's live D
J: Ok, but everything needs an ID and a curator, right ? What should it be for those ?
D:For the curator we can get the information from the file citace220Picture_timeStamp.ace that i put on tazendra under draciti. I will probably be the curator of the picture objects once I have associated the anatomy terms. For most of those the curator is Wen, but for many it says acedb or citace, which are not valid curators. You can tell me to assign them all to Wen (if Wen's okay with it, for Interactions she told Xiaodong to use Xiaodong as the curator) or be more specific.. -- J J put me as curator whenever it says acedb or citace, I have to go through them anyway.. :) D Well, in that case, which timestamp should I key the curator off of ? It would really be easier to just assign curator as a rule, but if you want to match the timestamp, I'd need to know which, because there's only one curator for the whole entry, but there are three curators in the timestamp. One after the Picture ID, one after the Expr_pattern tag, and one after the Expr_pattern data. This is not a real entry, but if it said this, which one should be the curator ? J put always me as curator also for existing picture objects, in this way it will be easier - D
Picture : "1_#9f7.jpeg" -O "2001-09-10_21:07:30_acedb"
Expr_pattern -O "2002-04-01_18:48:26_citace" "Expr55" -O "2002-04-01_18:48:26_wen"
J:Anything else that should be assigned, like curation status or anything else ? -- J
D: At the moment they should also be flagged as No dump since I have to manually curate them
Thanks again J and please let me know if I missed something fundamental...;) can well be...:P Feliz fin de semana! Dani
Assign no_dump, curator, source, expr_pattern, reference, and that's it, correct ? -- J Correct And assign IDs starting from 1 and going forward WBPicture0000000001. Thanks!! D
Files for old Picture_objects
The total No of old picture objects belonging to the picture class is 7228. See citace220Picture.ace file in this dir /Users/danielaraciti/Desktop/Wormbase/Files from Wen/PictureObjects
We have deleted the no dump flag for all the old picture objects on March 22nd so that the old objects will be dumped but we did not update the history table.
main directory on canopus: /home/daniela/sort_old_pictures
Juancarlos put the old picture objects for sorting on canopus here: /home/daniela/sort_old_pictures/5367
March 23rd. J converted the png files -coming from chronograms- into jpg files. Also the postgres tables were converted accordingly. The picture source file was converted in jpg. the same is true for the pictures having a jpeg extension.
convert file.png file.jpg convert_failed_png_to_jpg.sh convert_png_to_jpg.sh
J checked that all the pictures present in postgres till pgid 7228 (included) have a corresponding file on canopus.
the location on canopus for the old picture was originally:
5367 picture files are stored in Canopus here: daniela@canopus:/usr/local/wormbase/website-shared-files/html/images-website-classic/expression/patterns
2233 pictures (related to chronograms) are in this dir daniela@canopus:/usr/local/wormbase/website-shared-files/html/images-website-classic/expression/localizome$
Adding up 5367 and 2233 we should have 7599 pics. Canopus has a total of 7595 pictures files -> 4 pics discrepancy could be other files
3 files are on tazendra and NOT on Canopus. Check the list in compare_tazendra_to_canopus.outfile in canopus in the following dir: /home/daniela/sort_old_pictures
10_#9f7.jpg in tazendra, not on canopus WBPaper00001469
Expr3071.jpg in tazendra, not on canopus WBPaper00024532. This picture is a cartoon of the worm for the old renderings. It is also located on canopus on this location: /usr/local/wormbase/images_for_raymond/expression/assembled/Expr3071.png
Expr3072_3073.jpg in tazendra, not on canopus WBPaper00024532
Expr3071.jpg and Expr3072_3073.jpg correspond to the publication WBPaper00024532. The pictures were annotated directly by Daniela form the original publication. The 2 entries for the old pictures (pgid 4460 and 4461) could be removed for 2 reasons: A- the 2 riginal jpg files are not on canopus (one of the 2 was retrieved in another location and happened to be a cartoon, see above) B- the annotation of the pictures coming from the paper is in picture objects 7459, 7460, 7461. pgid 4460 and 4461 removed from Postgres on March 24th
/usr/local/wormbase/images_for_raymond/expression/assembled/Expr3071.png
Acknowledgments for old picture objects were set by default to persons and not to papers, we had therefore removed the reference from the reference field and put it as brief citation in the remark field. This is because the standard rule for acknowledgment for entries that have papers will follow the paper pipeline and search for a mapping for the publication. We have fixed all those entries either manually or programmatically. The excel file with details on that is /Users/danielaraciti/Desktop/Wormbase/Old_Pictures. We had as well added the contact and Person name to all the entries till 7228
For the old pictures named .png and .jpeg Juancarlos used Imagemagik to convert them in jpg in a way that all the picture file names are in jpg extension. This will make the downstream processing more uniform. All the chronograms were .png and were all converted in jpg. As well all the files named jpeg were converted in jpg. directory : /home/acedb/draciti/oa_picture_fixes/20110324_source_jpg/
script : fix_source_jpg.pl
To Do
Parse Expr.ace file into .obo file for term info and for paper term info. On tazendra at /home/acedb/draciti/Expr_pattern/ExprWS221.ace
We already did this for generegulation OA -- J
Legend
* text : text * bigtext : like longtext, but makes the text box expand when you click in it so you can see everything you've written * dropdown : few values * ontology : controlled vocabulary (tell me where they come from) * multiontology / multidropdown : (allows multiple values) * toggle : on / off, yes/no etc.
Daniela TODO when live on Tazendra to fit in old Picture data
- c09f9_3_larv.jpg (PGID5645) has many expr Expr2060 Expr2006 done 110329 DR
- Expr3072_3073.jpg has many expr Expr3072 Expr3073 Objects deleted --see above for details 110329 DR
- f59b2_13_can.jpg has many expr Expr19 Expr8 done 110329 DR
- f59b2_13_head.jpg has many expr Expr19 Expr8 done 110329 DR
- zc84_3_all.jpg (PGID7078) has many expr Expr25 Expr26 done 110329 DR
- zc84_3_vnc.jpg has many expr Expr25 Expr26 done 110329 DR
And Expr19 has a paper, but Expr8 does not. Expr25 has, Expr26 does not. done 110329 DR Assign Manually both Expr_patterns to the pictures
Add Description for 2 entries: Expr3071.png and Expr3072_3073.png --see above for details 110329 DR
Another thing that would be good is to set up a chronjob for updating the Mappings.txt file.
Final .ace file should be dumped as
Picture : WBPicture0000000001
Description "Figure Legend: A. ..... B. ..... C. .... D .....""
Name "WBPaper12345678_journal.pbio.0020352.g006_B" -- I need to have in this field the "_"
Cropped_from "journal.pbio.0020352.g006"
Remark "Some remark"
Expr_pattern "Expr1234"
Anatomy "WBbt:0005175"
Anatomy "WBbt:0003681"
Cellular_component "GO:123456"
Template "WormBase thanks the journal <Journal_URL> for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from <Journal_URL>, <Article_URL>. Copyright <Publication_year> with permission from <Publisher_URL>." -- take this from Template Text from Mappings.txt
Publication_year "2004"
Article_URL WBPaper00024505_URL id 0020352 -- Reference_URL id accession number.
Journal_URL "PLoS Biology" -- Take Full Journal Name from the Mappings.txt file on tazendra
Publisher_URL "PLoS" -- Take it from Publisher_name from Mappings.txt file
Reference "WBPaper00024505"
Contact "WBPersonID"
Database : WBPaper00024505_URL -- Reference_URL
Name "Ding M et al. (2008) PLoS One \"The cell signaling adaptor protein EPS-8 is essential for C. elegans epidermal ....\"" -- take this from Brief citation from Paper model NB There are "" that have to be escaped with backslash \ otherwise the .ace file is not reading in fine. Brief_citation name coming from new module at /home/postgres/work/citace_upload/papers/get_brief_citation.pm -- J
URL_constructor "http:\/\/www.plosbiology.org\/article\/info:doi%2F10.1371%2Fjournal.pbio.%S" -- take this from Article_URL from Mappings.txt
Database : "PLoS Biology" Take it from Full Journal Name from Mappings.txt
Name "PLoS Biology" Take it from Full Journal Name from Mappings.txt
URL_constructor "http:\/\/www.plosbiology.org\/" -- take this from Journal_URL from Mappings.txt
Database : PLoS -- Publisher_name in Mappings.txt
Name "PLoS" -- Publisher_name in Mappings.txt
URL_constructor "http:\/\/www.plos.org\/" take this from Publisher_URL from Mappings.txt
.ace dumper at mangolassi at /home/acedb/draciti/oa_picture_ace_dumper/
(actually at /home/postgres/work/citace_upload/picture/ and symlinked here)
called dump_picture_ace.pl
generates pictures.ace and pictures.err (errorfile, always look at this even if it's usually empty) -- J
Daniela, once you are in Mangolassi
cd acedb/draciti/oa_picture_ace_dumper
give command
./dump_picture_ace.pl
it generates 2 files: picture.ace and pictures.err
Sample curation results for a parental image when parental ≠ cropped
Name "WBPicture0000000001"
Reference "WBPaper00024505"
Descritpion "(A) A portion of the promoter sequence of K07C11.4 from C. elegans (bottom) aligned with its ortholog from C. briggsae (top). Boxed regions show conserved predicted PHA-4 binding sites and Early-1 and Early-2 elements. Site-directed mutations that disrupt Early-1 and Early-2 (“E2 + E1 Mut”) are shown below their respective wild-type (“E2 + E1 WT”) sequence from K07C11.4. (B–E) Confocal images of mid-stage embryos expressing GFP under the control of the wild-type K07C11.4 promoter (B) or promoters with a mutation in Early-1 (C), Early-2 (D), or both Early-1 and Early-2 (E). Percentages are the fraction of transgenic embryos expressing GFP; the remainder of embryos do not express GFP. (F) Expression of the wild-type K07C11.4 reporter in a subset of somatic gonad cells in an L4 animal (arrowheads). (G) Mutation of the Early-1 element eliminates gonadal expression but does not strongly affect expression in other tissues, such as intestinal cells (arrows). Dashed lines indicate the outline of the developing pharynx."
Source "journal.pbio.0020352.g006"
Expr_pattern "Expr3097"
Remark "N/A"
Cellular_component "N/A"
Anatomy_term "N/A"
Acknowledgments "WormBase wishes to thank the journal Genetics for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from Genetics, 166:151-60, Chen J, Li XJ, Greenwald I. sel-7, a positive regulator of lin-12 activity, encodes a novel nuclear protein in Caenorhabditis elegans. Copyright (2004) with permission from the Genetics Society of America."
Sample curation results for a daughter image
Name "WBPicture0000000002"
Reference "WBPaper00024505"
Descritpion "Confocal images of mid-stage embryos expressing GFP under the control of the wild-type K07C11.4 promoter"
Source "journal.pbio.0020352.g006_B"
Cropped_from "journal.pbio.0020352.g006"
Expr_pattern "Expr3097"
Remark "N/A"
Cellular_component "GO_term"
Anatomy_term "WBbt:0003681 pharynx"
Acknowledgments "WormBase wishes to thank the journal Genetics for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from Genetics, 166:151-60, Chen J, Li XJ, Greenwald I. sel-7, a positive regulator of lin-12 activity, encodes a novel nuclear protein in Caenorhabditis elegans. Copyright (2004) with permission from the Genetics Society of America."
Sample curation results for a parental image when parental = cropped
Name "WBPicture0000000003"
Reference "WBPaper00024876"
Descritpion "Expression Pattern of rom-1::nls::gfp Expression pattern of the zhIs5[rom-1::nls::gfprom-1::] transcriptional reporter during vulval development. Images on the left (A, C, E, G, and I) show the corresponding Nomarski pictures with the arrows pointing at the Pn.p cell nuclei and the arrowhead indicating the position of the AC nucleus. (B) A mid L2 larva before vulval induction with uniform rom-1::nls::gfp expression in all the Pn.p cells. (D) An early L3 larva in which rom-1::nls::gfp expression was decreased in all VPCs except P6.p (see text for a quantification of the expression pattern). Note that the nuclei of hyp7 and the Pn.p cells that had fused to hyp7 displayed strong rom-1::nls::gfp expression (P1.p, P2.p, P3.p and P9.p in the example shown). (F) A mid to late L3 larva in which P6.p had generated four descendants. Expression of rom-1::nls::gfp occurred only in the 3° descendants of P.4.p and P8.p after they fused to hyp7. (H) An L4 larva during vulval invagination. No rom-1::nls::gfp was detectable in the 1° and 2° descendants of P5.p, P6.p, and P7.p, but the AC and the surrounding uterine cells displayed strong rom-1::nls::gfp expression. (K) A late L2 to early L3 larva following the ablation of the precursors of the somatic gonad. No up-regulation of rom-1::nls::gfp in P5.p, P6.p, or P7.p was observed. The scale bar in (K) is 10 μm."
Source "journal.pbio.0020334.g003"
Expr_pattern "Expr3457"
Remark "N/A"
Cellular_component "WBbt:0004017 Cell"
Anatomy_term "N/A"
Acknowledgments "WormBase wishes to thank the journal Genetics for permission to reproduce figures from this article. Please note that this material may be protected by copyright. Reprinted from Genetics, 166:151-60, Chen J, Li XJ, Greenwald I. sel-7, a positive regulator of lin-12 activity, encodes a novel nuclear protein in Caenorhabditis elegans. Copyright (2004) with permission from the Genetics Society of America."
Model testing
On the temrinal
Go to acedb_good folder
then
./xace /Users/danielaraciti/Desktop/Wormbase/ACEDB/ts
you have opened the empty database ready for testing.
then you can -> EDIT -> Read models -> yes -> continue
NOTES
For testing the model, the path is reading the models.wrm file that is in ts -> wspec -> models.wrm
Note that all the files that go to acedb should be plain text, so if you want to modify the models.wrm file you should convert it into plain text (in text edit -> format make plain text)
when you test a new model you should first go into ts/wspec, replace the old model with the new one, save and launch again the ts
If you get an error that says error line No 123: Edit -> Find -> Select line and you go to the selected line
At the moment the file I am playing around with is models.wrm. the original file was named modelsoriginal.wrm
In case you need to add back Variation to the ?Picture data model add the following tag in ?Variation model: Picture ?Picture XREF Variation #Evidence
and add in the ?Picture model the following Variation ?Variation XREF Picture
same true for RNAi and Transgene
CHANGES TO THE MODELS.WRM FILE
?Picture Description Text // not modified
Name UNIQUE Text // Added in ?Picture
Crop Crop_picture ?Picture XREF Cropped_from // added in ?Picture
Cropped_from ?Picture XREF Crop_picture //added in ?Picture
Pick me to call Text Text // not modified
Expr_pattern ?Expr_pattern XREF Picture // not modified
RNAi ?RNAi XREF Picture // deleted from ?Picture and deleted the XREF to Picture in RNAi class
Variation ?Variation XREF Picture // deleted from ?Picture and deleted the XREF to Picture in Variation class
Transgene ?Transgene XREF Picture // deleted from ?Picture and deleted the XREF to Picture in Transgene class
Remark ?Text #Evidence // not modified
Cellular_component ?GO_term XREF Picture //added in ?Picture and added "Picture ?Picture XREF Cellular_component" in ?GO_term
Anatomy_term ?Anatomy_term XREF Picture //added in ?Picture and added Picture ?Picture XREF Anatomy_term in ?Anatomy_term
Acknowledgments Template Text // added in ?Picture
Journal_name Text // added in ?Picture
Publication_year Text // added in ?Picture
Article_URL ?Database ?Database_field ?Accession_number // added in ?Picture
Publisher_URL ?Database ?Database_field ?Accession_number // added in ?Picture
Person_name Text // added in ?Picture
Reference ?Paper XREF Picture // added in ?Picture and added Picture ?Picture XREF Reference in ?Paper
Delete Expression
in the folder OA Picture fixes there is a list of picture objects that are parental images to which I asked J to mass delete the XREF to Expr_pattern in a way that the Expr_pattern page would not have linked the parental images. Location: /home/acedb/draciti/oa_picture_fixes/20110321_delete_expression
file name: pic_exprpattern.pg
that's the backup of the 135 entries to delete. Done DR 14.04.2011
go to http://tazendra.caltech.edu/~postgres/cgi-bin/referenceform.cgi
To check the list of parental images which have a cropped from (use this to see which picture object should have the Expr_pattern XREF removed) SELECT * FROM pic_exprpattern WHERE pic_exprpattern IS NOT NULL AND joinkey IN (SELECT joinkey FROM pic_name WHERE pic_name IN (SELECT pic_croppedfrom FROM pic_croppedfrom) );
Identifying and retrieving pictures associated with Expression Pattern
- 1. Use the curation status form to get a list of papers that do have curatable images. This includes images for which a paper has been curated for gene expression and for which we hold copyright permission.
http://tazendra.caltech.edu/~postgres/cgi-bin/curation_status.cgi
select curator and click on Curation statistics option Page. Select Pictures. It will retrieve a list of papers that have been curated for Expression_pattern and could potentially have expression images. As of now (Aug 2013) it displays all the papers, please note that we might not have permission to reproduce images for all of them. In order to get a list of papers that contain images and are curatable (for which we hold permission) one should
In this directory on tazendra : /home/acedb/draciti/picture_curatable/ Journals foe which we have permission are in this file (add journals here if we obtain more): journal_with_permission And run this script : picture_flagged_permission.pl What it does is get the papers from this URL : http://tazendra.caltech.edu/~postgres/cgi-bin/curation_status.cgi?action=listCurationStatisticsPapersPage&select_curator=two1&listDatatype=picture&method=any%20pos%20ncur&checkbox_cfp=on&checkbox_afp=on&checkbox_svm=on and filter against the pap_journal table through the list of journal_with_permission You can run it like : ./picture_flagged_permission.pl > out And look at the file 'out'
- 2. Go to Textpresso.dev and run the image extraction pipeline using the WBPaperID list gathered in step #1 --using Arun's script-- this is the location on Textpresso-dev:
/data1/Users/liyuling/Curator_related/expression_pattern/scripts/
Command: ./02legends_images_pdftohtml.pl "pdf_dir" "output_dir" And one more command to package the output_dir into tar.gz tar -zcvf output_dir.tar.gz output_dir
- 3. Once you have the folders with the Pictures
Contacting publishers to request permission on a paper-by-paper basis is too time consuming. Along the lines of other mods (zfin) we will retrieve only images from journals that are open access or granted us blanket permission. We could revise this pipeline in a year and re-negotiate with major publishers (e.g. AAAS).
Flagging a paper negative for pictures
- go to the curation status form: http://tazendra.caltech.edu/~postgres/cgi-bin/curation_status.cgi
- Select you name and click on 'Specific paper page'
- Enter the WBPaperID, e.g.: WBPaper00001234
- Select Picture
- Hit 'Get results'
- In the 'New result' box select validated negative
Identifying and retrieving pictures associated with topics
December 2014: All topic diagram pictures will be put in OA regardless of permission. We will set manually the no dump toggle for images that do not have permission. the dumper will give an error message in case there is a topic for a WBpaper that does not have permission
- process objects only dump if there's a paper and the paper has a journal
- the script looks at a file listing papers with permission, which is located here: /home/acedb/draciti/picture_curatable/journal_with_permission
- all the rest of the pipeline is the same as for Expression images: gets Mappings Journal/Publisher from /home/acedb/draciti/oa_picture_ace_dumper/Mappings.txt and URL accession numbers from /home/acedb/draciti/picture_source/picture_source
the following query will give you a list of the WBPapers flagged relevant for a specific topic. Modify only the WBbioprID
SELECT DISTINCT(pro_paper) FROM pro_paper WHERE joinkey IN (SELECT joinkey FROM pro_process WHERE pro_process ~ 'WBbiopr:00000046') AND joinkey IN (SELECT joinkey FROM pro_topicpaperstatus WHERE pro_topicpaperstatus = 'relevant') ORDER BY pro_paper;
In this directory on tazendra : /home/acedb/draciti/picture_curatable/
run like
./picture_flagged_permission.pl file
to use
http://tazendra.caltech.edu/~acedb/draciti/papers_with_topic_pictures.txt
edit file at
/home/acedb/public_html/draciti/papers_with_topic_pictures.txt
the script will get the list of WBPaerIDs that are in the URL and compare it with the list of journals for which we have permission. We are using it to identify images of processes but it can be used for other classes as well.
NB: in the directory there are 2 files:
journal_with_permission journal_with_permission_expr
- journal_with_permission lists the open access papers (the ones we consider curatable for topic)
if you run the script ./picture_flagged_permission.pl file you will get a list of the papers curatable for topic pictures
- journal_with_permission_expr lists the papers for which we have blanket permission to display expression pattern images -including Elsevier
to get the list of papers you just run the script ./picture_flagged_permission.pl
whenever you need to add/delete a paper you just do it on the list and the changes will automatically reflect in OA
Permissions
Below is a list of Journals for which we hold blanket permissions or are open access. A more detailed list is on Lario/Publishers/Permissions in Curation status.xlsx
Mol Biol Cell Genome Biol J Biol Neural Dev BMC Biol BMC Cell Biol BMC Dev Biol BMC Evol Biol BMC Genet BMC Genomics BMC Mol Biol BMC Neurosci BMC Physiol Genetics G3 COMP FUNCT GENOM PLoS Biol PLoS Comput Biol PLoS Genet PLoS Pathog PLoS One J Cell Biol J Gen Physiol Biochem Biophys Res Commun Biochim Biophys Acta Cancer Cell Cell Cell Host & Microbe Cell Metab Cell Signal Cell Stem Cell Chemistry and Biology Curr Biol Dev Cell Exp Cell Res Gene Gene Expr Patterns Genomics Immunity J Mol Biol J Struct Biol Mech Dev Mol Cell Neuron Structure Trends Parasitol
We obtained an 'ad hoc' permission for the following journals
Biochem J Biochemistry Biol Cell Biol Chem Biotechniques Comp Funct Genomics DNA Cell Biol Development Dis Model Mech Endocrinology FASEB J Folia Biol (Praha) Genes Dev Genome Res J Am Soc Nephrol J Biol Chem J Cell Sci J Exp Biol J Histochem Cytochem J Neurosci PNAS Proc Natl Acad Sci U S A RNA Sci Signal Science
Mappings for the new_fetch.pl script
location of the script /home/acedb/draciti/picture_curatable/Pictures_next_level/new_fetch.pl
The script worked well for BMC, Genetics, JCell Biol but did not initially work for science direct journals. We troubleshooted and got it to work but then again science direct changed the url for the high quality picture files. This is not mantainable in the long run as they keep changing the urls so we have retrieved the images with Arun's script (for 240 papers). In the future we should probably just use Arun's script as it is more reliable and agnostic on url changes.
Publisher Fetch_pictures journals AMER SOC CELL BIOLOGY Mol_Biol_Cell/Molbiolcell Mol Biol Cell BIOMED CENTRAL LTD BMC/BMC Genome Biol BIOMED CENTRAL LTD BMC/BMC J Biol BIOMED CENTRAL LTD BMC/BMC Neural Dev BIOMED CENTRAL LTD BMC/BMC BMC Biol BIOMED CENTRAL LTD BMC/BMC BMC Cell Biol BIOMED CENTRAL LTD BMC/BMC BMC Dev Biol BIOMED CENTRAL LTD BMC/BMC BMC Evol Biol BIOMED CENTRAL LTD BMC/BMC BMC Genet BIOMED CENTRAL LTD BMC/BMC BMC Genomics BIOMED CENTRAL LTD BMC/BMC BMC Mol Biol BIOMED CENTRAL LTD BMC/BMC BMC Neurosci BIOMED CENTRAL LTD BMC/BMC BMC Physiol GENETICS SOC AM Genetics/Genetics Genetics GENETICS SOC AM Genetics/Genetics G3 PLoS PLoS/PLoS PLoS Biol PLoS PLoS/PLoS PLoS Comput Biol PLoS PLoS/PLoS PLoS Genet PLoS PLoS/PLoS PLoS Pathog PLoS PLoS/PLoS PLoS One ROCKEFELLER UNIV PRESS JCellBiol/JCellBiol J Cell Biol Elsevier BiochemBiophysResCommun/BiochemBiophysResCommun Biochem Biophys Res Commun Elsevier Cell/Cell Cell Elsevier CurrBiol/CurrBiol Curr Biol Elsevier DevCell/DevCell Dev Cell Elsevier Gene/Gene Gene Elsevier JMolBiol/JMolBiol J Mol Biol Elsevier MechDev/MechDev Mech Dev Elsevier MolCell/MolCell Mol Cell Elsevier Neuron Neuron For the following papers I will retrieve pics with Arun's script (too few to be worth a separate script) HINDAWI PUBLISHING CORPORATION COMP FUNCT GENOM ROCKEFELLER UNIV PRESS J Gen Physiol Elsevier Biochim Biophys Acta Elsevier Cancer Cell Elsevier Cell Host & Microbe Elsevier Cell Metab Elsevier Cell Signal Elsevier Cell Stem Cell Elsevier Chemistry and Biology Elsevier Exp Cell Res Elsevier Gene Expr Patterns Elsevier Genomics Elsevier Immunity Elsevier J Struct Biol Elsevier Structure Elsevier Trends Parasitol
Correspondence with Publishers about Permission is on Lario/Publishers, arranged by alphabetical order
Automating Publisher Permission request
Total number of papers with attached Expr_pattern object in WS221: 2406. We have contacted 48 publishers in order to request permission for image display into Wormbase. We have been able to obtain permission to reproduce images for 1162 papers (26 major publishers) and we are negotiating with 7 publishers to obtain permission for additional 1182 papers (On May 12th 2011). 3 publishers either did not accept the request or asked for a fee to reproduce the figures (38 papers). 13 publishers did not get back (24 papers).
To obtain permission for newly published images we want to set up an automated system so that each image will be requested during the curation process. We will set up a pilot with National Academy of Sciences since they agreed to receive requests as they come up. We will extend the system to other publishers once developed.
Pictures will be curated during normal curation process (e.g. with Expr_patterns). Once a month a cronjob will read into OA and send an e-mail to the publisher requesting permission. Sample e-mail:
Dear <Publisher_name> we are requesting permission to reproduce the following material into Wormbase. - volume number, issue number, and issue date - article title - authors' names - Figure/table number We will reproduce the material into Wormbase, a non profit educational website. Intended audience: researchers and students. Please click here if you are accepting to grant us permission or here if you do not agree. Thank you for your collaboration. Daniela Raciti, PhD California Institute of Technology Curator, www.Wormbase.org Mailing Address: Division of Biology Mail Code 156-29 California Institute of Technology 1200 E. California Blvd. Pasadena, CA 91125 Office Phone: (626) 395-8613 E-mail: draciti@caltech.edu
If this set-up will turn out to be good, we will move on and contact 6 additional major publishers. In turn we can extend the pipeline to smaller publishers.
Fetch Pictures
Contact Dev Biol again for permission (211 papers)
Juancarlos' script fetch_pictures.pl (/Users/danielaraciti/Desktop/Fetchpictures) retrieves images from jorunals. So far, images were retrieved from:
- Development (The Company of Biologists) 281 papers (181 able to fetch, 100 required Arun's script)
- JBC (Amer Soc Biochemistry Molecular Biology) 109 papers (99 able to fetch, 10 required Arun's script)
- Genes Dev (COLD SPRING HARBOR LAB PRESS) 97 papers (70 able to fetch, 27 required Arun's script)
- Mol Biol Cell (AMER SOC CELL BIOLOGY) 104 papers (99 able to fetch, 5 required Arun's script)
- Journal of Cell Science (Company of Biologists) 55 papers (42 able to fetch, 13 required Arun's script)
- J Cell Biol (Rockefeller university press) 74 papers (65 able to fetch, 9 required Arun's script)
- Cell (Elsevier Cell Press) 104 papers (79 able to fetch, 25 required Arun's script)
- Neuron (Elsevier Cell Press) 58 papers (56 able to fetch, 2 required Arun's script)
- Biochem Biophys Res Commun (Elsevier) 33 papers (23 able to fetch, 10 required Arun's script)
- Gene (Elsevier) 23 papers (12 able to fetch, 6 required Arun's script, 5 missing)
- J Mol Biol (Elsevier) 46 papers (37 able to fetch, 9 required Arun's script)
- Mech Dev (Elsevier) 21 papers (19 able to fetch, 1 required Arun's script, 1 missing)
For the following journals (Elsevier publisher) we have received permission but the number of papers is too low for being worth to write a fetching script so we will retrieve figures using Arun's script (36 papers total) search done May 1st 2012:
- Biochim Biophys Acta
- Cancer Cell
- Cell Host & Microbe
- Cell Metab
- Cell Signal
- Cell Stem Cell
- Chemistry and Biology
- Exp Cell Res
- Gene Expr Patterns
- Genomics
- Immunity
- J Struct Biol
- Structure
- Trends Parasitol
For the following journals (Elsevier) we did not obtain permission but the number of papers is too low. Only FEBS Lett would be worth a try
- Arch Biochem Biophys 1
- Biochimie 1
- Brain Res 2
- Mol Brain Res 1
- Chemical Physics 1
- DNA Repair (Amst) 5
- Eur J Cell Biol 2
- Exp Gerontol 1
- FEBS Lett 19
- Free Radic Biol Med 1
- Int J Parasit 7
- J Neurosci Methods 2
- Mech Ageing Dev 6
- Mol Biochem Parasitol 3
- Mol Cell Biol Res Commun 1
- Mol Cell Neurosci 2
- Molecular & Biochemical Parasitology 1
- Mutat Res 1
- Neurobiol Aging 1
- Neuropharmacology 1
- Neurosci Lett 3
- Neuroscience 1
- Parkinsonism Relat Disord 1
- Toxicol Lett 1
Textpresso mining pipeline
We are developing pipeline in order to achieve automatic picture retrieval. The first trial was done with Development journal as 281 papers contained gene expression pattern objects. Juancarlos wrote a script to fetch pictures directly from the journal web-page so that the quality of the pictures will be the highest possible. On lario: /Users/danielaraciti/Desktop/Fetchpictures/fetch_pictures.pl. the script is extracting also the url for each paper and that url will be automatically dumped in the .ace file without need to insert it in the OA.
Out of 281 papers, 181 returned a positive result and all the figures and figure legends were extracted. For the remaining 100 papers Arun set up a pipeline for figure and figure caption mining form the pdfs stored in Wormbase. Briefly, every figure in the pdf is extracted and associated with a figure legend in a way that the figure name figure legend name will correspond. Subsequently the pdf is scanned again and converted into html with a pdf to html converter and the figures are again extracted with a higher quality. The first round of extraction is not sufficient to achieve a decent quality. In addition, a rule for flagging positive matches was developed, in a way that every figure caption contains a positive or negative flag.
The set of 100 papers that went through Arun script were manually curated. 96 of them were taken for further analysis (1 paper did not contain pictures associated with Expr_pattern and I need to get back to the remaining 3 papers). The goal was to calculate the script recall and precision.
Recall = 119/143 = 0.832167832167832
Precision = 119/193 = 0.616580310880829
Importing Wormatlas dataset
Wormatlas contains 1999 pictures of gene expression. They all come from the Moerman large expression study. Zeynep agreed to let us display the pictures in the picture page. Juancarlos and I queried postgres and we already had 1734 out of 1999 pictures. We have been able to download the remaining 265 (264 because one object was empty). The 264 images don't have a direct link to an Expression pattern object. What we want to do is to create new picture object is to script the info that are in the pic name. e.g. first digits before the _ are correspnding to the locus (e.g. AH6.11_BC12595_GFP_a-2_1.jpg). In order to autopopulate the Expression Pattern field in the picture OA we cannot browse directly postgres tables because that large scale study had not been imported into OA (still on Citace minus). What we should do is to find the ExprID directly on ACeDB. Whenever there are multiple expression pattern objects corresponding to the same gene, we should pick the one related to this publication: Hunt-Newbury R et al. (2007) PLoS Biol. High-throughput in vivo analysis of gene expression in Caenorhabditis ...
Daniela and Juancarlos worked on getting the mapping sequence-name -> gene -> Expr-object to import them into OA. All the files are in the Wormatlas folder on tazendra /home/acedb/draciti/worm_atlas.
The parsing into Picture OA will be as follows:
- pgid from 9116 on
- pic_name from 9116 on
- pic_contact WBPerson427
- pic_source name of the file e.g. B0024.14b_BC13961_GFP_a-1_1.jpg
- pic_exprpattern Exprxxxx
- pic_remark Hunt-Newbury R et al. (2007) PLoS Biol. High-throughput in vivo analysis of gene expression in Caenorhabditis ...
- pic_person WBPerson427
- pic_curator WBPerson12028
Script to populate: /home/postgres/work/pgpopulation/pic_picture/20110919_wormatlas/populate_wormatlas.pl
NB: sometimes one gene has associated more than one Expression pattern (see tazendra /home/acedb/draciti/worm_atlas/out2), e.g. picture 01E11.7_BC10244_GFPV_a-1_1bi.jpg, WBGene00006508 Expr6402,Expr6403. However in OA the picture is associated only to Expr6402 as the strain associated to 6402 is BC10244 while the strain associated to 6403 is BC14499.
Itai Yanai large scale import -WBPaper00041190
In order to display pictures of expression time course we needed to generate expression objects. The objects (Expression and Picture) will be deleted once Wen will finish curating microarray for all species described in the paper and once we will have in place a way to generate images of expression on the fly - data will be retrieved directly from SPELL.
For now Daniela and Juancarlos have generated 2 .ace files, one for pictures and one for expression. the files are on CitaceMinus. The files are called expr_pattern_Yanai.ace and pictures_Yanai.ace
Expression pattern and Picture objects were given high numbers so when the new display system will be in place those could be deleted without affecting anything in OA.
Expression objects go from Expr1010178 to Expr1029229
Picture objects go from WBPicture0001011201 to WBPicture0001030251
there are 19052 objects for each class.
GeneAce old objects
in May 2015
- 1000 more picture objects in WS compared to citace or citace minus (ftp://ftp.sanger.ac.uk/pub2/wormbase/STAFF/pad/Daniela/bad_pictures.ace)
- Paul Davis found them coming from GeneAce
- Have "pick_me_to_call" tag
- Represent images from Ian Hope's lab (old Sylvia Martinelli objects)
- Objects are redundant with what we already have and will be deleted
Daniela and Juancarlos working on checking redundancies and importing the descriptions form Sylvia Martinelli to the pic_description field in OA files here: /home/postgres/work/pgpopulation/pic_picture/20150528_compare_sylvia_data
matching lists the files that are in OA out of 1001 only 13 are not:
NOMATCH UL#155C2.1.jpg NOMATCH UL#155C2.2.jpg NOMATCH UL#155C2.3.jpg NOMATCH UL#64A1.1.jpg NOMATCH UL#64A1.2.jpg NOMATCH r107_1_emb.jpg NOMATCH r107_1_gnp.jpg NOMATCH r107_1_lar.jpg NOMATCH r11a5.2_adult.jpg NOMATCH r11a5.2_head.jpg NOMATCH r11a5.2_tail.jpg NOMATCH r11a5.2_tail2.jpg NOMATCH r11a5.2_vulv.jpg
Daniela checked them the original names (e.g. UL#155C2.1.jpg) were converted into UL#155C2_1.jpg -underscore instead of full stop- for file_name reasons. We have them all but NOMATCH r107_1_emb.jpg NOMATCH r107_1_gnp.jpg NOMATCH r107_1_lar.jpg
the file out is the output of the descriptions that will go populate the Picture OA in the pic_description field
Obsolete Summary Pipeline for picture handling
- Pictures are saved and organized in folders in Lario (Daniela's computer) as described above. The folder name will be the WBPaperID or the WBPersonID
- Generate 600/folder containing 600x600 full view with Photoshop image processor
- Generate 200/ folder containing original files and 200x200 thumbnails with Thumbs Up
- Go to /Users/danielaraciti/Desktop/Canopus/Pictures and run script ./mergeToOICR.pl The script that juancarlos wrote will # get all subfolders from the 200/ folder, # move them to the OICR folder, # get all subfolders for the 600/ folder, # get each jpg in those subfolders, # for each of those create a new name for it that names has _600 in it before the .jpg
- Check for errors (should automatically display errors if any). 200/ will be empty and 600/ will contain a number of empty folders. Delete them.
- Go to /Users/danielaraciti/Desktop/Canopus/Pictures and scp the file that was generated - picture_source- to tazendra scp picture_source acedb@tazendra.caltech.edu:/home/acedb/draciti/picture_source/
- rsync the file to Canopus rsync --progress --delete-after -a /Users/danielaraciti/Desktop/Canopus/Pictures/OICR/ daniela@canopus.caltech.edu:OICR/
Now you should see the .jpg file names and a link to the .txt file for the figure legend in the term info (takes it from picture_source) and OICR should have access to Canopus to get the actual files
To update the term info display into picture OA on tazendra:
Go to /home/acedb/draciti/picture_source/populate_obo_data_pic_picturesource.pl
this script updates directly the term info display into the OA> You have now links to the new pictures.
OICR should take the pictures from:
canopus.caltech.edu:/usr/local/wormbase/pictures/picture_object
Obsolete Pipeline for picture handling
Picture are saved on Lario (draciti's computer) as JPG files in directories organized after the WBPaperID. The original files will be converted in 200x200 pixels thumbnails and 600x600 pixels full view according the following:
Generating 200x200px thumbnails
Thumbnails are generated using the freeware "ThumbsUp" (v4.4) a simple, drag-and-drop based utility to create thumbnails for a bunch of pictures and supports all image formats of Mac OS X and QuickTime (including PDF documents)<ref>http://www.macupdate.com/info.php/id/11898/thumbsup</ref>
Trials for automation have been done with Photoshop (automated image processor) and MacOSX (Automator -> creation of Thumbnail images). With Photoshop automator is NOT possible to save the thumbnails in the same folder. With MacOSX Automator is not possible to create thumbnails larger than 128px. ThumbsUp allows generation of 200x200 in the same folder where the original files are.
Therefore, in this folder -called 200- we have the reference file and the 200x200 thumbnail
The file name for thumbnails is the same as the original picture with a _200 suffix
Generating 600x600px full view
600x600 images are generated with photoshop (scripts -> image processor) and stored in a separate folder called 600. The architecture of the sub-folders is the same as the original. It is not possible to generate the 600x600 with Thumbs up because it will not maintain the folder architecture if saved in a separate folder.
Merging ref, 200x200, and 600x600 pictures
We need to programmatically rename all the pictures that are in draciti/600/ folder. Then we will take the files from /600 and /200 and put them all together in a new folder called Merged (run script ./put_stuff_together.pl - Daniela modify this with the correct name). The script takes files from 600/ and 200/ and puts it together in draciti/merged
Then we rsync draciti/merged to Canopus. Once this is done I move 600/ and 200/ to draciti/done The next time I have a batch of pictures I recreate 600/ and 200/ and rerun ./put_stuff_together.pl
As a general comment. For the beginning the pipeline for picture curation will be done publisher by publisher. Once a batch of pictures is downloaded from PLoS we will do a batch conversion. Later on when we will work with single papers we will try to make the picture conversion part of the curation.
Canopus scripts
the script is called transfer_from_Pictures_to_good.pl
and is located here on canopus
/home/daniela
the script transfers jpg files from $dirSource to $dirDest if subdirectory+file have been curated in picture OA. 2012 10 18. The script is
use File::Copy;
is a non standard module that needs to be installed
here is where pics are coming from
my $dirSource = '/home/daniela/all_pictures';
this is where pics are going to
my $dirDest = '/home/daniela/OICR/Pictures/';
this hash stores pics that have been curated. the structure of the %curated hash maps to the the paper or person, then the file
my %curated;
the following line requests to have the Yanai file
my $yanai_file = 'pictures_Yanai.ace';
opens the Yanai file. Since Yanai's pics are not in OA we need to read from a flat file. Its capturing those and is mapping them to the curated Hash
- $curated{"WBPerson4037"}{$file}++
- %curated -> WBPerson4037 -> filename
in the Yanai file the filename looks like this:
Name "WBGene00003442.jpg"
Then we are querying postgres for the picture source and the papers for the picture source
- SELECT pic_source.pic_source, pic_paper.pic_paper, pic_paper.joinkey FROM pic_source, pic_paper WHERE pic_source.joinkey = pic_paper.joinkey
we get the filename, paper, pgid and we put it in the curated hash %curated -> paper -> filename
my ($filename, $paper, $pgid) = $row =~ m/<TD>(.*?)<\/TD>/g;
now we are doing the same query again
- SELECT pic_source.pic_source, pic_person.pic_person, pic_person.joinkey FROM pic_source, pic_person WHERE pic_source.joinkey = pic_person.joinkey
but instead of querying for papers we query for persons
and doing the same things
we get the filename, paper, pgid and we put it in the curated hash %curated -> person -> filename
in this query there could be multiple people and this is the line doing it
my (@persons) = $allpersons =~ m/(WBPerson\d+)/g;
and it goes into
%curated -> each_person -> filename
my %source;
my @source = <${dirSource}/*>;
foreach my $source (@source) {
if (-d $source) {
my (@subsource) = <${source}/*.jpg>;
my (@subsource2) = <${source}/*.JPG>;
foreach my $subsource (@subsource, @subsource2) {
if (-f $subsource) {
my (@stuff) = split/\//, $subsource;
my $file = pop @stuff;
my $dir = pop @stuff;
next unless ($curated{$dir}{$file});
# print "SOURCE $file D $dir E\n";
$source{$dir}{$file}++; } } } }
it reads into the source dir, it looks at each thing that exists there. It only looks at directories. Opens the dir and looks for .jpg or .JPG and if any of those are files -as opposed to dir or symlinks- it gets the file and the dir and puts into curated directory file %curated -> directory -> file and this works because the name of the directories are named WBPaper######## or WBPerson########
the same thing happens for the destination hash for the destination source
my %dest;
my @dest = <${dirDest}/*>;
foreach my $dest (@dest) {
if (-d $dest) {
my (@subdest) = <${dest}/*.jpg>;
my (@subdest2) = <${dest}/*.JPG>;
foreach my $subdest (@subdest, @subdest2) {
if (-f $subdest) {
my (@stuff) = split/\//, $subdest;
my $file = pop @stuff;
my $dir = pop @stuff;
$dest{$dir}{$file}++; } } } }
foreach my $dir (sort keys %source) {
unless ($dest{$dir}) {
my $newDir = $dirDest . '/' . $dir;
unless (-e $newDir) {
mkdir $newDir, 0755;
print "mkdir $newDir\n"; } }
foreach my $file (sort keys %{ $source{$dir} }) {
unless ($dest{$dir}{$file}) {
copy("${dirSource}/${dir}/$file", "${dirDest}/${dir}/$file");
print "copy ${dirSource}/${dir}/$file ${dirDest}/${dir}/$file\n"; }
} # foreach my $file (sort keys %{ $source{$dir} })
} # foreach my $dir (sort keys %source)
if it exists in the source hash but it does not exist in the dest hash, it creates a directory and make writable-executable It goes through each file and if it exists in the source but not in the destination is copying the file over
foreach my $dir (sort keys %dest) {
my $deleteDir = 0;
unless ($source{$dir}) { $deleteDir = $dirDest . '/' . $dir; }
foreach my $file (sort keys %{ $dest{$dir} }) {
unless ($source{$dir}{$file}) {
unlink("${dirDest}/${dir}/$file");
print "rm ${dirDest}/${dir}/$file\n"; }
} # foreach my $file (sort keys %{ $dest{$dir} })
if ($deleteDir) {
rmdir $deleteDir;
print "rmdir $deleteDir\n"; }
} # foreach my $dir (sort keys %dest)
If it exists in the destination but not in the source get rid of the files and delete them and deletes the directory
Virtual Worm images
We will import Chris' renderings generated with Blender into a widget on the Anatomy pages.
The renderings are located here:
http://canopus.caltech.edu/virtualworm/Anatomy_Images/
Juancarlos wrote a script to compare the names of the jpgs in the Virtual Worm directories above to the anatomy ontology obo file.
The script is on tazendra here:
cd azurebrd/work/parsings/daniela/20160624_virtualworm
output in 'out'
Obsolete PMID retrieval for pictures
This is how papers containing expression data were retrieved for picture curation. This pipeline became obsolete after the curation status form went live ~Jan 2013
- Secure shell to spica
- go to dir Daniela/GetPMID
- modify the file JournalList.txt with the name of the journals you want to retrieve the papers containing expression data
- run the script ./getJournalPMID.sh
- the output with the list of papers containing Expr dtata is in JournalPaperWithExpr.ace and contains the WBPaperID and the PMID
- transfer the JournalPaperWithExpr.ace to your local machine, rename it after the Journal name and put it in a folder called after the journal name. E.g. enerate a folder called Science and rename the .ace file science.ace.
- GREP the PMIDs identifiers e.g.: GREP PMID Science.ace > PMID. In this way you have a file containing all the PMIDs
- run Yuling script to generate a PMID list that could be copied in pubmed. Goto Desktop/Scripts/SCRIPT PMID and then run the command "./grab_pmid.pl input output"
- Transfer the output file into the "Science" folder
- Copy the PMID list and paste it in the Pubmed search box
- Click search and send to file choosing xml format. Save it in the "Science" folder as Science.xml