Gene Interaction
'links to relevant pages
Caltech documentation
Interaction
OA link
Contents
Interaction Curation
Pipeline
semi-automatic curation with textpresso extracted sentences
- There are 2138 sentences (actually 2133 sentences) in the sourcefile /home/postgres/work/pgpopulation/genegeneinteraction/20091002-xiaodong/ggi_20091002
- paper starts at WBPaper00028425, ends at WBPaper00035225
- .ace dumper at /home/acedb/xiaodong/gene_gene_interaction/dump_ggi_ace.pl
- go to the directory and do: ./dump_ggi_ace.pl > some_file.ace
- Populate textpresso data in tazendra OA: done on 20110110 -X**
- cd to directory on tazendra: /home/acedb/xiaodong/textpresso_ggi
- mkdir directionay_name (eg 20110106)
- cd directory_name (eg 20110106)
- get Arun's result file (35225-35725.txt in the directory)
- run script: ./populate_textpresso_ggi_to_OA.pl 20110106/35225-35725.txt WBPerson4793 > 20110106/35225-35725.pg (with first argument file_name as input file, and second argument WBPersonID, then output file)
- after running, '20110106/35225-35725.pg' should be in '20111106' directory.
upload Gary and Chris RNAi-based interaction objects into OA
- Reading file created by Igor's script into aceDB
- you can use the empty database by ssh -X citpub@spica.caltech.edu
- then cd CitaceMirror
- then type 'ts' to launch an empty acedb
- Dumping no-worry .ace file
- Then parse into OA
- scp ace file to same directory as below
- ssh acedb@tazendra.caltech.edu (directory xiaodong/interaction_ace_parsing)
- To run : ./parse_ace_interaction_oa.pl gary_RNAi.ace WBPerson1823
- where gary_RNAi.ace is the <inputfile>,the first argument, and WBPerson# is the second argument.
dump .ace file from OA for upload
- on tazendra: /home/acedb/xiaodong/oa_interactions_dumper/use_package.pl
- run script by calling: ./use_package.pl
- error file will be spitted out in the same directory after every run. inform curators to check the errors.
Interaction Model
?Interaction Evidence #Evidence
Interactor ?Gene XREF Interaction #Interactor_info
Interaction_type Genetic #Interaction_info
Regulatory #Interaction_info
No_interaction #Interaction_info
Predicted_interaction #Interaction_info
Physical_interaction #Interaction_info
Suppression #Interaction_info
Enhancement #Interaction_info
Synthetic #Interaction_info
Epistasis #Interaction_info
Mutual_enhancement #Interaction_info
Mutual_suppression #Interaction_info
Confidence Confidence_level UNIQUE Float
P_value UNIQUE Float
Log_likelihood_score UNIQUE Float
Paper ?Paper XREF Interaction
DB_info Database ?Database ?Database_field ?Accession_number
Remark ?Text #Evidence
#Interactor_info
Variation ?Variation XREF Interactor
Transgene ?Transgene XREF Interactor
Remark ?Text #Evidence //info about the reagents that the model can't capture goes here (e.g. co_suppression, RNA_reagent, etc.)
#Interaction_info
Interaction_RNAi ?RNAi XREF Interaction
Effector ?Gene //master, upstream
Effected ?Gene //subject, downstream
Non_directional ?Gene //e.g. synthetic interactions - Igor
Interaction_phenotype ?Phenotype XREF Interaction
Confidence Confidence_level UNIQUE Float
P_value UNIQUE Float
#Interaction_type
Genetic //directional and non_directional
Physical_interaction
Regulation
No_interaction
Synthetic//non_directional
Epistasis
Enhancement
Suppression
Predicted //addition for WeiWei, non_direactional
Mutual_enhancement//non_directional
Mutual_suprression//non_directional
///////////////////////////////////////////////////////////////////////////////////
Gene_gene Interaction OA
OA interface
- Tab 1
- PGID
- Interaction ID
- no interaction ID is generate by clicking on 'new'. when 'duplicate', the ID from old entry will be in the field, but need to be deleted in order to get an new ID.
- Interaction ID will be assigned by cronjob daily at 4 am.
- If you mistakenly make a typo and assign a correct ID's value to some other ID, you will _not_ be able to bring it back (because it's an ontology) without going to postgres directly and editing the int_name and int_name_hst tables by pgid (in postgres called joinkey). You'll have to note the pgid and then manually change it in postgres.
- Non_directional
- toggle OFF (default), means interaction is directional. there is effected/effector parties involve in the object.
- toggle ON (color change to red by click) means interaction is non_directional
- Interaction Type//dropdown list with 11 types showing in .ace template
- Effected Gene //autocomplete WBGene, multiontology, corresponding to interactor in .ace file. order does not matter when dump to interactors
- Effected Variation //WBGene, WBVar, multiontology, autocomplete on Variation, store in separate lines ->.ace, Interactor "WBGene" Variation "WBVar"
- Effected Transgene_Name //ontology, autocomplete transgene object name, eg iaIs3.
- Effected Transgene_Gene // multi-ontology, autocomplete WBGene->.ace, Interactor "WBGene" Transgene "id". In case of multi genes, WBGene is followed by same transgene id. One wbgene for each .ace line ? Make sure you really want it this way, we can go with product/promoter if that's what you want, just make sure it's what you want. It matters having extra fields and scrolling and so forth. You'll see when the text fields become multi-ontology and ontology.
- Effected Other Type //dropdown list of 'Chemicals' and 'Transgene'
- Effected Other //free text field now, however, when entering chemicals make sure to enter common names followed by mesh IDs in parenthesis for later ontologinization.
- two fields above will be dumped in remark field.
- Effector Gene //autocomplete WBGene, multiontology. corresponding to interactor in .ace file. order does not matter when dump to interactors
- Effector Variation //WBGene, WBVar, autocomplete multiontology on variation, store in separate lines ->.ace, Interactor "WBGene" Variation "WBVar". use name server to map variation to gene, or the file Karen gave you to map variation to gene for variation OA.
- Effector Transgene_Name //autocomplete name, ontology
- Effectot Transgene_Gene // autocomplete WBGene, multi-ontology, ->.ace, Interactor "WBGene" Transgene "id". In case of multi genes, WBGene is followed by same transgene id.
- Effector Other Type //dropdown list of 'Chemicals' and 'Transgene'
- Effector Other //free text field
- two fields above will be dumped in remark field.
Note: Gene, Variation, and Transgene_Gene all refer to different genes. There is no pairing problem.
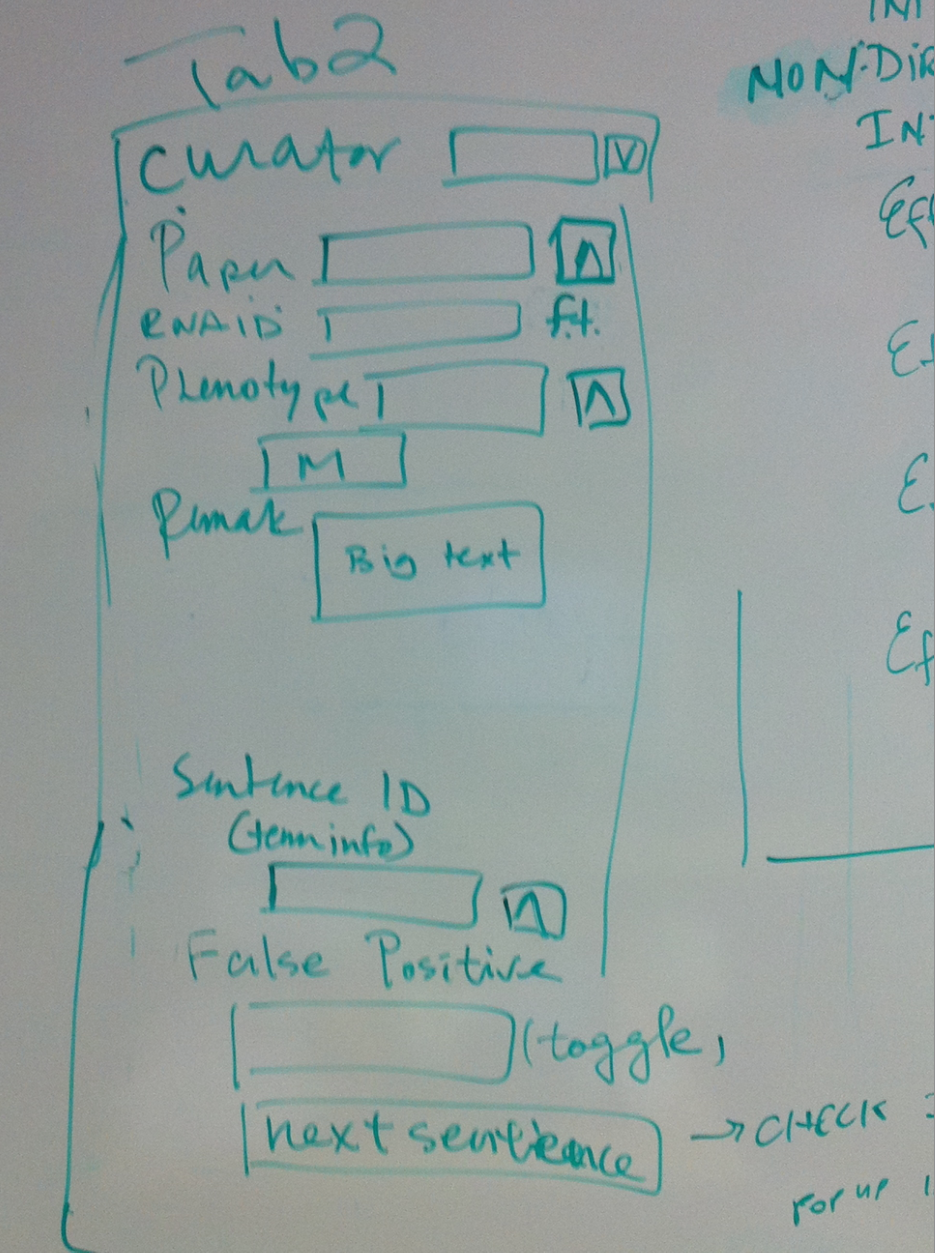
- Tab 2
- Curator//dropdown list
- Paper//ontology
- RNAi ID//free text fiel
- Phenotype//multiontology
- Remark//big text
- Sentence ID//sentence shows in term info
- False Positive//toggle, will not give an id or no dump if the sentence is false positive, containing no interaction info
.ace template:
- Interaction : ""
- Interactor "WBGene" Variation ""
- Interactor "WBGene" Transgene ""
- Interactor "WBGene"
- Interaction_type Genetic Effector ""
- Interaction_type Genetic Effected ""
- Interaction_type Genetic Non_directional ""
- Interaction_type Genetic Interaction_RNAi ""
- Interaction_type Genetic Interaction_phenotype ""
- Interaction_type Regulatory Effector ""
- Interaction_type Regulatory Effected ""
- Interaction_type Regulatory Non_directional ""
- Interaction_type Regulatory Interaction_RNAi ""
- Interaction_type Regulatory Interaction_phenotype ""
- Interaction_type No_interaction Effector ""
- Interaction_type No_interaction Effected ""
- Interaction_type No_interaction Non_directional ""
- Interaction_type No_interaction Interaction_RNAi ""
- Interaction_type No_interaction Interaction_phenotype ""
- Interaction_type Predicted_interaction Non_directional ""
- Interaction_type Predicted_interaction Interaction_RNAi ""
- Interaction_type Predicted_interaction Interaction_phenotype ""
- Interaction_type Physical_interaction Effector ""
- Interaction_type Physical_interaction Effected ""
- Interaction_type Physical_interaction Interaction_RNAi ""
- Interaction_type Physical_interaction Interaction_phenotype ""
- Interaction_type Suppression Effector ""
- Interaction_type Suppression Effected ""
- Interaction_type Suppression Interaction_RNAi ""
- Interaction_type Suppression Interaction_phenotype ""
- Interaction_type Enhancement Effector ""
- Interaction_type Enhancement Effected ""
- Interaction_type Enhancement Interaction_RNAi ""
- Interaction_type Enhancement Interaction_phenotype ""
- Interaction_type Synthetic Non_directional ""
- Interaction_type Synthetic Interaction_RNAi ""
- Interaction_type Synthetic Interaction_phenotype ""
- Interaction_type Epistasis Effector ""
- Interaction_type Epistasis Effected ""
- Interaction_type Epistasis Interaction_RNAi ""
- Interaction_type Epistasis Interaction_phenotype ""
- Interaction_type Mutual_enhancement Non_directional ""
- Interaction_type Mutual_enhancement Interaction_RNAi ""
- Interaction_type Mutual_enhancement Interaction_phenotype ""
- Interaction_type Mutual_suppression Non_directional ""
- Interaction_type Mutual_suppression Interaction_RNAi ""
- Interaction_type Mutual_suppression Interaction_phenotype ""
- Paper ""
- Remark ""
interaction objects source file
- there are 9242 interaction objects dumped from WS220 on Monday, 10/01/2010
- Juancarlos's parse results from this file:
/home/postgres/work/pgpopulation/interaction/20101004_xiaodong_start/out
There are two interactions in postgres, but not the .ace file : In postgres, no ace WBInteraction0008637 In postgres, no ace WBInteraction0008638//will be OA
There are 1290 interactions in .ace file not in postgres (so I imagine these are what we should read in ?)//these are RNAi based interaction objects, we want to include them in OA
There are >40000 interactions that have a ticket and are in neither .ace nor postgres.//these 398,619 interactions are from two large scale papers
Also there are interaction data in postgres without an interaction ID//will be assigned id from WBInteraction0500001.
Some Notes for gene_gene_interaction
- two large scale papers
- WBPaper00027155 (Weiwei's science paper) has 23128 objects, starting from WBInteraction0008637 and ending at WBInteraction0050578 (blank ids in between)
- WBPaper00031465 (Lee's Nature Genetics paper) has 375491 objects, starting from WBInteraction 0100001, ending at WBInteraction0475491 (which is the largest WBInteraction id in acedb)
- directories on tazendra related to gene_gene_interaction (home/acedb/xiaodong)
- assigning_interaction_ids
- textpresso_ggi
- interaction_ace_parsing
- oa_interactions_dumper