Difference between revisions of "Gene Interaction"
| Line 79: | Line 79: | ||
**Non_directional //toggle on (default), off (color change by click) means effected/effector on. | **Non_directional //toggle on (default), off (color change by click) means effected/effector on. | ||
**Interaction Type//dropdown list with 11 types showing in .ace template | **Interaction Type//dropdown list with 11 types showing in .ace template | ||
| − | **'''Effected Gene | + | **'''Effected Gene //WBGene, multiontology, corresponding to interactor in .ace file. order does not matter when dump to interactors''' |
| + | **'''Effected Variation //WBGene, WBVar, multiontology, store in separate lines ->.ace, Interactor "WBGene" Variation "WBVar". use name server to map variation to gene, or the file Karen gave you to map variation to gene for variation OA. ''' | ||
**'''Effected Transgene_Name //ontology''' | **'''Effected Transgene_Name //ontology''' | ||
| − | **'''Effected | + | **'''Effected Transgene_Gene // multi-ontology, ->.ace, Interactor "WBGene" Transgene "id". In case of multi genes, WBGene is followed by same transgene id.''' |
| − | **'''Effector Gene | + | **'''Effector Gene //WBGene, multiontology, corresponding to interactor in .ace file. order does not matter when dump to interactors''' |
| − | **'''Effector Transgene_Name //ontology''' | + | **'''Effector Variation //WBGene, WBVar, multiontology, store in separate lines ->.ace, Interactor "WBGene" Variation "WBVar". use name server to map variation to gene, or the file Karen gave you to map variation to gene for variation OA. '''**'''Effector Transgene_Name //ontology''' |
| − | **'''Effectot | + | **'''Effectot Transgene_Gene // multi-ontology, ->.ace, Interactor "WBGene" Transgene "id". In case of multi genes, WBGene is followed by same transgene id.''' |
<br> | <br> | ||
Revision as of 23:36, 3 November 2010
'links to relevant pages
Caltech documentation
Interaction
Contents
Interaction Curation
Pipeline
semi-automatic curation with textpresso extracted sentences
- There are 2138 sentences (actually 2133 sentences) in the sourcefile /home/postgres/work/pgpopulation/genegeneinteraction/20091002-xiaodong/ggi_20091002
- paper starts at WBPaper00028425, ends at WBPaper00035225
- .ace dumper at /home/acedb/xiaodong/gene_gene_interaction/dump_ggi_ace.pl
- go to the directory and do: ./dump_ggi_ace.pl > some_file.ace
Interaction Model
?Interaction Evidence #Evidence
Interactor ?Gene XREF Interaction #Interactor_info
Interaction_type Genetic #Interaction_info
Regulatory #Interaction_info
No_interaction #Interaction_info
Predicted_interaction #Interaction_info
Physical_interaction #Interaction_info
Suppression #Interaction_info
Enhancement #Interaction_info
Synthetic #Interaction_info
Epistasis #Interaction_info
Mutual_enhancement #Interaction_info
Mutual_suppression #Interaction_info
Confidence Confidence_level UNIQUE Float
P_value UNIQUE Float
Log_likelihood_score UNIQUE Float
Paper ?Paper XREF Interaction
DB_info Database ?Database ?Database_field ?Accession_number
Remark ?Text #Evidence
#Interactor_info
Variation ?Variation XREF Interactor
Transgene ?Transgene XREF Interactor
Remark ?Text #Evidence //info about the reagents that the model can't capture goes here (e.g. co_suppression, RNA_reagent, etc.)
#Interaction_info
Interaction_RNAi ?RNAi XREF Interaction
Effector ?Gene //master, upstream
Effected ?Gene //subject, downstream
Non_directional ?Gene //e.g. synthetic interactions - Igor
Interaction_phenotype ?Phenotype XREF Interaction
Confidence Confidence_level UNIQUE Float
P_value UNIQUE Float
#Interaction_type
Genetic //directional and non_directional
Physical_interaction
Regulation
No_interaction
Synthetic//non_directional
Epistasis
Enhancement
Suppression
Predicted //addition for WeiWei, non_direactional
Mutual_enhancement//non_directional
Mutual_suprression//non_directional
///////////////////////////////////////////////////////////////////////////////////
Gene_gene Interaction OA
OA interface
- Tab 1
- PGID
- Interaction ID//generate automatically per entry
- Non_directional //toggle on (default), off (color change by click) means effected/effector on.
- Interaction Type//dropdown list with 11 types showing in .ace template
- Effected Gene //WBGene, multiontology, corresponding to interactor in .ace file. order does not matter when dump to interactors
- Effected Variation //WBGene, WBVar, multiontology, store in separate lines ->.ace, Interactor "WBGene" Variation "WBVar". use name server to map variation to gene, or the file Karen gave you to map variation to gene for variation OA.
- Effected Transgene_Name //ontology
- Effected Transgene_Gene // multi-ontology, ->.ace, Interactor "WBGene" Transgene "id". In case of multi genes, WBGene is followed by same transgene id.
- Effector Gene //WBGene, multiontology, corresponding to interactor in .ace file. order does not matter when dump to interactors
- Effector Variation //WBGene, WBVar, multiontology, store in separate lines ->.ace, Interactor "WBGene" Variation "WBVar". use name server to map variation to gene, or the file Karen gave you to map variation to gene for variation OA. **Effector Transgene_Name //ontology
- Effectot Transgene_Gene // multi-ontology, ->.ace, Interactor "WBGene" Transgene "id". In case of multi genes, WBGene is followed by same transgene id.
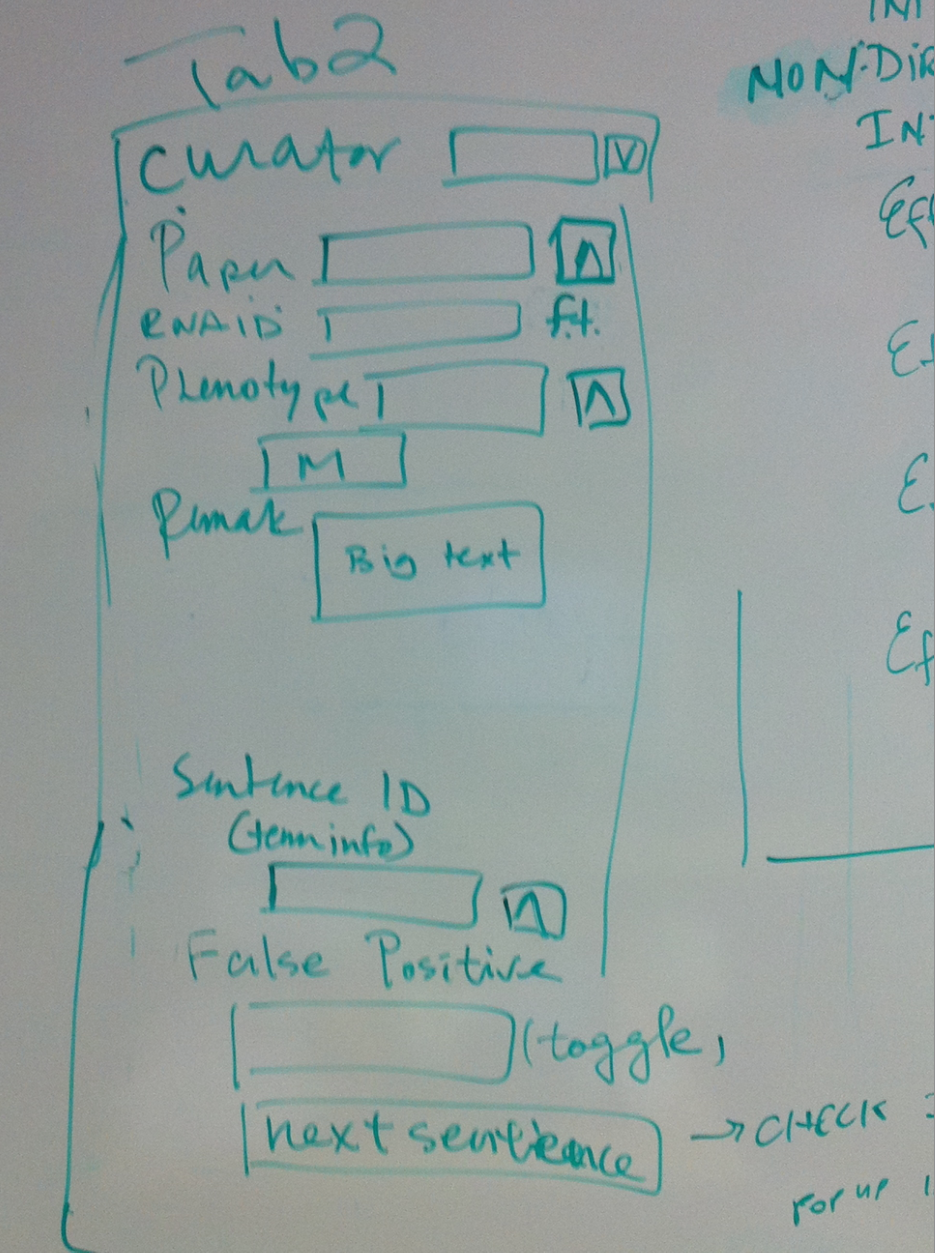
- Tab 2
- Curator//dropdown list
- Paper//ontology
- RNAi ID//free text fiel
- Phenotype//multiontology
- Remark//big text
- Sentence ID//sentence shows in term info
- False Positive//toggle, will not give an id or no dump if the sentence is false positive, containing no interaction info
- next sentence button
.ace template:
- Interaction : ""
- Interactor "WBGene" Variation ""
- Interactor "WBGene" Transgene ""
- Interactor "WBGene"
- Interaction_type Genetic Effector ""
- Interaction_type Genetic Effected ""
- Interaction_type Genetic Non_directional ""
- Interaction_type Genetic Interaction_RNAi ""
- Interaction_type Genetic Interaction_phenotype ""
- Interaction_type Regulatory Effector ""
- Interaction_type Regulatory Effected ""
- Interaction_type Regulatory Non_directional ""
- Interaction_type Regulatory Interaction_RNAi ""
- Interaction_type Regulatory Interaction_phenotype ""
- Interaction_type No_interaction Effector ""
- Interaction_type No_interaction Effected ""
- Interaction_type No_interaction Non_directional ""
- Interaction_type No_interaction Interaction_RNAi ""
- Interaction_type No_interaction Interaction_phenotype ""
- Interaction_type Predicted_interaction Non_directional ""
- Interaction_type Predicted_interaction Interaction_RNAi ""
- Interaction_type Predicted_interaction Interaction_phenotype ""
- Interaction_type Physical_interaction Effector ""
- Interaction_type Physical_interaction Effected ""
- Interaction_type Physical_interaction Interaction_RNAi ""
- Interaction_type Physical_interaction Interaction_phenotype ""
- Interaction_type Suppression Effector ""
- Interaction_type Suppression Effected ""
- Interaction_type Suppression Interaction_RNAi ""
- Interaction_type Suppression Interaction_phenotype ""
- Interaction_type Enhancement Effector ""
- Interaction_type Enhancement Effected ""
- Interaction_type Enhancement Interaction_RNAi ""
- Interaction_type Enhancement Interaction_phenotype ""
- Interaction_type Synthetic Non_directional ""
- Interaction_type Synthetic Interaction_RNAi ""
- Interaction_type Synthetic Interaction_phenotype ""
- Interaction_type Epistasis Effector ""
- Interaction_type Epistasis Effected ""
- Interaction_type Epistasis Interaction_RNAi ""
- Interaction_type Epistasis Interaction_phenotype ""
- Interaction_type Mutual_enhancement Non_directional ""
- Interaction_type Mutual_enhancement Interaction_RNAi ""
- Interaction_type Mutual_enhancement Interaction_phenotype ""
- Interaction_type Mutual_suppression Non_directional ""
- Interaction_type Mutual_suppression Interaction_RNAi ""
- Interaction_type Mutual_suppression Interaction_phenotype ""
- Paper ""
- Remark ""
interaction objects source file
- there are 9242 interaction objects dumped from WS220 on Monday, 10/01/2010
- Juancarlos's parse results from this file:
/home/postgres/work/pgpopulation/interaction/20101004_xiaodong_start/out
There are two interactions in postgres, but not the .ace file : In postgres, no ace WBInteraction0008637 In postgres, no ace WBInteraction0008638//will be OA
There are 1290 interactions in .ace file not in postgres (so I imagine these are what we should read in ?)//these are RNAi based interaction objects, we want to include them in OA
There are >40000 interactions that have a ticket and are in neither .ace nor postgres.//these 398,619 interactions are from two large scale papers
Also there are interaction data in postgres without an interaction ID//will be assigned id from WBInteraction0500001.
Some Notes for gene_gene_interaction
- two large scale papers
- WBPaper00027155 (Weiwei's science paper) has 23128 objects, starting from WBInteraction0008637 and ending at WBInteraction0050578 (blank ids in between)
- WBPaper00031465 (Lee's Nature Genetics paper) has 375491 objects, starting from WBInteraction 0100001, ending at WBInteraction0475491 (which is the largest WBInteraction id in acedb)