Difference between revisions of "Co-op Documentation"
| Line 1: | Line 1: | ||
| + | ==Current Tasks== | ||
| + | Tasks to be completed. | ||
| + | *Refining Interactions widget: | ||
| + | **Details: | ||
| + | ***If the number of interactions is too high, only show the minimal table (hide Cytoscape) and add "Show More" option(AJAX). | ||
| + | ***Otherwise, show all interactions(nearby interactions included) and Cytoscape plugin | ||
| + | **Progress: | ||
| + | ***Looking into BLASTP field in Homology Widget on Protein page that uses similar mechanism | ||
| + | *Gene page alterations | ||
| + | **Details: | ||
| + | ***Make corrections to Gene page interface | ||
| + | **Progress: | ||
| + | ***Pending consultation from curators. Teleconference: 11:30 am EST Wednesday, January 18th, 2012 | ||
| + | Possible Tasks: | ||
| + | Additional tasks that require further investigation. | ||
| + | *Loading/processing large amounts of data | ||
| + | **Details: | ||
| + | ***Likely approach will be to change the database used from ACeDB to NoSQL/CouchDB | ||
| + | **Progress: | ||
| + | ***[https://github.com/aduong/ace2couch Ace2Couch Scripts] for migrating data | ||
| + | ***[https://github.com/aduong/AceCouch AceCouch Perl API] | ||
| + | ==Getting Started== | ||
| + | ===Members/Contacts=== | ||
| + | *Todd Harris | ||
| + | *Abigail Cabunoc | ||
| + | *Lincoln Stein | ||
| + | *Quang Trinh | ||
| + | ===Meetings=== | ||
| + | *WormBase OICR Developers Teleconference | ||
| + | **Mondays, 3:00PM | ||
| + | **Phone-in 1-800-747-5150 id: 6738514 | ||
| + | *WormBase OICR Developers Teleconference with Lincoln | ||
| + | **Wednesdays, 4:00PM EST | ||
| + | **Lincoln's office | ||
| + | *WormBase International Groups Teleconference | ||
| + | **Every other Thursday, 11:30AM | ||
| + | **Lincoln's Office | ||
| + | *Group Meeting | ||
| + | **Fridays, 3:00PM | ||
==Tools== | ==Tools== | ||
The following are some of the tools with which many major aspects of WormBase are developed. | The following are some of the tools with which many major aspects of WormBase are developed. | ||
===Perl=== | ===Perl=== | ||
| − | There is significant documentation on getting started with Perl. One starting point is | + | There is significant documentation on getting started with Perl. One starting point is [http://perlmonks.org/?node=Tutorials PerlMonks] |
| − | |||
===Catalyst=== | ===Catalyst=== | ||
Catalyst is the web development framework used to develop WormBase. To get started, read | Catalyst is the web development framework used to develop WormBase. To get started, read | ||
| − | *The Definitive Guide to Catalyst | + | *The Definitive Guide to Catalyst (should be available on bookshelf) |
| + | * [http://search.cpan.org/~hkclark/Catalyst-Manual-5.9002/lib/Catalyst/Manual/About.pod CPAN - About Catalyst] | ||
===Git=== | ===Git=== | ||
Git is a version control system used for collaboration and backup in the development process. | Git is a version control system used for collaboration and backup in the development process. | ||
| Line 29: | Line 68: | ||
Note: pull is similar to using fetch + merge | Note: pull is similar to using fetch + merge | ||
===Other=== | ===Other=== | ||
| + | Some other tools that you should be aware of but may not be required to know/interact with include: | ||
| + | *JavaScript | ||
| + | *ACeDB | ||
| + | *MySQL | ||
| + | *Xapian | ||
| + | *GFF | ||
*Cytoscape Web | *Cytoscape Web | ||
| − | **[http://cytoscapeweb.cytoscape.org/tutorial Cytoscape] | + | **[http://cytoscapeweb.cytoscape.org/tutorial Cytoscape Tutorial] |
**Plugin generally used for pathway analysis | **Plugin generally used for pathway analysis | ||
**This plugin is used in the Interactions widget on the Gene page ([WormBase dir]/root/templates/classes/gene/interactions.tt2) | **This plugin is used in the Interactions widget on the Gene page ([WormBase dir]/root/templates/classes/gene/interactions.tt2) | ||
| Line 36: | Line 81: | ||
==General Concepts== | ==General Concepts== | ||
===Widget Data Loading=== | ===Widget Data Loading=== | ||
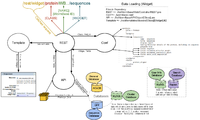
| − | [[File: | + | [[File:DataLoadingWidget1.png|200px|thumb|Widget Loading]] |
REST Controller: | REST Controller: | ||
#Catches internal url (/rest/widget/...) | #Catches internal url (/rest/widget/...) | ||
| Line 43: | Line 88: | ||
#Retrieves data for corresponding fields from API methods (/lib/WormBase/API/Object/[Class].pm) | #Retrieves data for corresponding fields from API methods (/lib/WormBase/API/Object/[Class].pm) | ||
#Sends data to the template to be used in rendering the widget (/root/templates/classes/[Class]/[Widget].tt2) | #Sends data to the template to be used in rendering the widget (/root/templates/classes/[Class]/[Widget].tt2) | ||
| − | + | ==Other Notes== | |
| − | ==Other== | + | *[WormBase dir]/root/templates/config/main contains useful macros such as get_url that may be used in templates |
| − | *[WormBase dir]/root/templates/config/main contains useful macros such as get_url | ||
Revision as of 18:56, 17 January 2012
Contents
Current Tasks
Tasks to be completed.
- Refining Interactions widget:
- Details:
- If the number of interactions is too high, only show the minimal table (hide Cytoscape) and add "Show More" option(AJAX).
- Otherwise, show all interactions(nearby interactions included) and Cytoscape plugin
- Progress:
- Looking into BLASTP field in Homology Widget on Protein page that uses similar mechanism
- Details:
- Gene page alterations
- Details:
- Make corrections to Gene page interface
- Progress:
- Pending consultation from curators. Teleconference: 11:30 am EST Wednesday, January 18th, 2012
- Details:
Possible Tasks: Additional tasks that require further investigation.
- Loading/processing large amounts of data
- Details:
- Likely approach will be to change the database used from ACeDB to NoSQL/CouchDB
- Progress:
- Ace2Couch Scripts for migrating data
- AceCouch Perl API
- Details:
Getting Started
Members/Contacts
- Todd Harris
- Abigail Cabunoc
- Lincoln Stein
- Quang Trinh
Meetings
- WormBase OICR Developers Teleconference
- Mondays, 3:00PM
- Phone-in 1-800-747-5150 id: 6738514
- WormBase OICR Developers Teleconference with Lincoln
- Wednesdays, 4:00PM EST
- Lincoln's office
- WormBase International Groups Teleconference
- Every other Thursday, 11:30AM
- Lincoln's Office
- Group Meeting
- Fridays, 3:00PM
Tools
The following are some of the tools with which many major aspects of WormBase are developed.
Perl
There is significant documentation on getting started with Perl. One starting point is PerlMonks
Catalyst
Catalyst is the web development framework used to develop WormBase. To get started, read
- The Definitive Guide to Catalyst (should be available on bookshelf)
- CPAN - About Catalyst
Git
Git is a version control system used for collaboration and backup in the development process.
- WormBase repository located at https://github.com/organizations/WormBase
- Common commands
- status, add, checkout, commit, push, pull, log, fetch, merge
- Usage example:
Given that we have modified two files a.txt and b.txt but do not wish to keep the changes made to b.txt
git status
git checkout b.txt
git add a.txt
git commit -m “Added change1 and change2 to a.txt”
git push
If we have did not have the most recent version, then we will run into an issue when trying to push. In this case we can:
git fetch
git merge
git push
Note: pull is similar to using fetch + merge
Other
Some other tools that you should be aware of but may not be required to know/interact with include:
- JavaScript
- ACeDB
- MySQL
- Xapian
- GFF
- Cytoscape Web
- Cytoscape Tutorial
- Plugin generally used for pathway analysis
- This plugin is used in the Interactions widget on the Gene page ([WormBase dir]/root/templates/classes/gene/interactions.tt2)
- Installed in [WormBase dir]/root/js/jquery/plugins/
General Concepts
Widget Data Loading
REST Controller:
- Catches internal url (/rest/widget/...)
- Determines the class and widget from the url
- From class and widget, determines which fields are required from the configuration file (wormbase.conf)
- Retrieves data for corresponding fields from API methods (/lib/WormBase/API/Object/[Class].pm)
- Sends data to the template to be used in rendering the widget (/root/templates/classes/[Class]/[Widget].tt2)
Other Notes
- [WormBase dir]/root/templates/config/main contains useful macros such as get_url that may be used in templates