|
|
| (108 intermediate revisions by 2 users not shown) |
| Line 1: |
Line 1: |
| − | =May 9 2017=
| |
| − | ==Agenda==
| |
| | | | |
| − | ===Arnold foundation===
| |
| − | * LoS for Coko
| |
| − | * Update from Paul (Mike Stebbins)
| |
| − | * Action plan
| |
| − | ** depending on IF and WHEN we will need to submit a proposal for Arnold:
| |
| − | - submit same proposal if U01 is turned down? will need to expand
| |
| − | - talk openly with Mike Stebbins and disclose that we are waiting on the NIH response and will tailor the Arnold proposal accordingly?
| |
| − | - if Coko gets the Arnold grant and we don’t, would they still build our infrastructure
| |
| | | | |
| − | ===Micropublication form updates===
| + | [[File:July2017.png|400px|thumb|center]] |
| − | * Added suggested reviewer field
| |
| − | * Added title field
| |
| − | * Added ISH
| |
| − | | |
| − | ===Potential submissions===
| |
| − | * Veena Prahlad (reviewer of a previous micropub) wants to submit smFISH data
| |
| − | | |
| − | ===Website views post blog===
| |
| − | * Still slight increase of website views
| |
| − | [[File:May9.png|400px|thumb|center]] | |
| − | | |
| − | ===Elsevier===
| |
| − | * will send examples as soon as we get data not shown/unpublished for their journals
| |
| − | | |
| − | =May 2 2017=
| |
| − | ==Agenda==
| |
| − | | |
| − | ===Elsevier===
| |
| − | * Gave us the OK to publish 'unpublished data' - do not specifically mention data not shown, which was in the original request
| |
| − | * they'd like to link back to our Database and propose 2 options:
| |
| − | | |
| − | ** 1. Banner linking as already established with Wormbase. Nonetheless, this is general (only Wormbase banner appearing on ScienceDirect) and if not already linked to Wormbase, the article will have to be re-supplied on our end, which means a very manual and time costly process.
| |
| − | ** 2. Evolving article feature. This feature will allow to host on our platform a specific PDF containing the additional information and links to it, that will be visualized as an additional pdf that readers can download when downloading the original article. See an example here: http://www.sciencedirect.com/science/article/pii/S1755436514000607 . It is still a manual process at the moment, but it does not involve re-supplying of the article.
| |
| − | | |
| − | * Shall we opt for the banner linking one?
| |
| − | | |
| − | ===Knudra/booklet===
| |
| − | * We met with Chris and Trisha (Knudra) and they will prepare a micropub submission, they will also reach out to their customers and see if they can supply one, too.
| |
| − | * they (and Matt Beaudet from Nemametrix) gave suggestions for the booklet, Matt strongly supports having PDFs versions of the articles in the booklet, even if hard to read. Chris from Knudra said they could provide magnifying glasses as swag
| |
| − | * Booklet link: https://docs.google.com/presentation/d/1MpxxYIxZ6eBXxWDmm8UuoHS244DBA9EAJb0AexFM1oM/edit#slide=id.p6
| |
| − | | |
| − | ===Blog post===
| |
| − | * We posted a call for expression data on the blog on Friday
| |
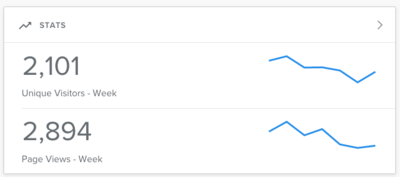
| − | * Since the post page views of micropublication biology have increased (from ~70-80 unique visitors per week to 115)
| |
| − | [[File:Stats.png|400px|thumb|center]]
| |
| − | | |
| − | ===micropub e-mail===
| |
| − | * Todd please correct misspelling in the mailing list
| |
| − | | |
| − | ===Other updates?===
| |
| − | * Jorgensen?
| |
| − | * Han will submit 2 micros by the end of next week
| |
| − | | |
| − | ==Minutes==
| |
| − | ===Elsevier===
| |
| − | * we will request to go with the banner linking option
| |
| − | | |
| − | ===Knudra/booklet===
| |
| − | * Better to have the pdf in one page other than spread over 2 pages
| |
| − | | |
| − | ===micropub e-mail===
| |
| − | * Todd fixed it
| |
| − | | |
| − | ===Other updates?===
| |
| − | * Todd contacted Jorgensen, no response yet
| |
| − | * Tim: plan for making the micropub easily found on the wormbase web homepage
| |
| − | ** Add a button on Wormbase called Micropublication: biology linking to website -between submit data and Parasite
| |
| − | * Arnold's foundation VP want to chat with Paul about micropubs
| |
| − | * Daniela will remove links to WBPapers that are not live in WormBase yet
| |
| − | * will meet next week, if no major updates will schedule in 2 weeks
| |
| − | | |
| − | =April 25 2017=
| |
| − | ==Agenda==
| |
| − | | |
| − | ===Data not shown===
| |
| − | 4 major publishers responded that is ok to publish data not shown or unpublished results:
| |
| − | * GSA
| |
| − | * The Company of Biologists (Publisher of Development, Disease Models and Mech, Journal of Cell Science, J Exp Biol)
| |
| − | * American Society of Cell Biology (Publisher of Mol Biol Cell)
| |
| − | * American Society of Microbiology (Publisher of Mol Cell Biol)
| |
| − | | |
| − | Waiting for Elsevier's final reply. They did not see anything wrong with it but they will double check with other colleagues.
| |
| − | | |
| − | It seems safe to continue to request data not shown unpublished results as micropubs for the journals above
| |
| − | | |
| − | ===IWM Booklet===
| |
| − | * booklet link: https://docs.google.com/presentation/d/1MpxxYIxZ6eBXxWDmm8UuoHS244DBA9EAJb0AexFM1oM/edit
| |
| − | | |
| − | Summary of Karen's discussion with Knudra/Nemametrix
| |
| − | | |
| − | * NemaMetrix would love to sponsor the printing of the micropub book up to $1K
| |
| − | * NemaMetrix and Knudra would love to have a "double "full page" ad in the booklet
| |
| − | * To maximize the impact of the micropub book distribution we would love to support you in following a standard 3,2,1 marketing launch (3 emails go out letting people know to be expecting the micropub book at the conference). These would be the strongest if they all came from WormBase but we can also support that with NemaMetrix and Knudra email lists
| |
| − | * We would be happy to add the 'micropub book' onto our daily giveaway raffle helping to drive traffic to your booth
| |
| − | * We would also be happy to participate in any follow-on activity such as surveys, etc.
| |
| − | | |
| − | * shall we have a raffle and give prizes to people that try to use the form? e.g. 2x 20$ amazon vouchers
| |
| − | * shall we have an extra swag to give away?
| |
| − | | |
| − | mini flashlight tool
| |
| − | | |
| − | https://www.anypromo.com/keychains-keylights/key-lights/mini-flashlight-tool-keychain-p672207
| |
| − | | |
| − | mints
| |
| − | | |
| − | https://www.4imprint.com/product/118346-24HR/Flip-Top-Dispenser-with-Sugar-Free-Mints-24-hr
| |
| − | | |
| − | * Todd, can the e-mail address micropublications@wormbase.org be used from users to contact us? I tried to send an e-mail from my caltech account but the group seem not be open to posting
| |
| − | | |
| − | ===Submissions===
| |
| − | | |
| − | * Han Wang (Paul's lab) will submit 2 micropubs, 1) novel good alleles for known genes that were recovered from forward genetic screens (allele phenotype info) 2) Gal4 drivers mapping information
| |
| − | | |
| − | * Karen and Daniela will have a meeting with Knudra this coming Thursday to get a submission from them
| |
| − | | |
| − | ==Minutes==
| |
| − | ===Data not shown===
| |
| − | * Focus on journals for which we have got the OK
| |
| − | * Will send urls back to publishers when 'data not shown' will be published on micropublication biology
| |
| − | | |
| − | ===IWM Booklet===
| |
| − | * Aim on publishing 1500 booklets
| |
| − | * Before the meeting we should advertise the project -and booklet- on WormBase: i)Blog post ii)Booklet Preview on the front page
| |
| − | * How we can make it valuable? We will have a daily raffle- 20$ Amazon voucher/day for people that try to submit
| |
| − | * for the raffle: 1) fake data -> 1 entry in the raffle 2) real data -> 100 entries in the raffle
| |
| − | * no extra swag
| |
| − | * we will have a banner
| |
| − | * Todd will make the micropublications@wormbase.org account public
| |
| − | | |
| − | ===Submissions===
| |
| − | * Todd will contact Erik M. Jorgensen for submission
| |
| − | * focus on getting few more examples out and focus on high value
| |
| − | | |
| − | =April 11 2017=
| |
| − | ==Agenda and Minutes==
| |
| − | * Knudra interested in micropubs. From Chris: "..new null alleles for unc-18 that we are characterizing and would love to understand what phenotypic and genotypic observation would be worthy of micro-publication?"
| |
| − | ** Karen will set up a meeting with Knudra to evaluate what kind of metadata they want to provide. Daniela reached out to Mary Ann to get an .ace example for engineered allele curation.
| |
| − |
| |
| − | * IWM booklet -mock-up here: [[https://docs.google.com/presentation/d/1MpxxYIxZ6eBXxWDmm8UuoHS244DBA9EAJb0AexFM1oM/edit#slide=id.p20]]
| |
| − | ** We need to add a slide on what micropubs are, maybe also a couple of blank pages (notepad style to allow people to take notes during the meeting). think of having an extra swag that we will give away if people come to the booth to give their e-mail address. Add an example of how to submit.
| |
| − | ** some ideas on swag here: https://www.4imprint.com/hot
| |
| − | ** Before the IWM we need to add a 'Micropublish' box on the WormBase homepage (linking out to the Micropublication:biology site), so it will be easy for users to get there.
| |
| − | * Will talk Friday April 14th with Ruth, Tracey about Data not shown
| |
| − | | |
| − | =April 4 2017=
| |
| − | ==Agenda and Minutes==
| |
| − | | |
| − | ===Tim's minipub===
| |
| − | * modified Tim's minipub to include micropubs
| |
| − | * trial version here: http://www.micropublicationbiology.org/minipub-kocsisova-et-al-2017.html
| |
| − | * expression micropubs here: http://www.micropublicationbiology.org/expression-data--tim.html
| |
| − | * phenotype micropubs here: http://www.micropublicationbiology.org/phenotype-data--tim.html
| |
| − | * if approved by Tim et al we could send out for review
| |
| − |
| |
| − | ** Tim will send a couple of corrections for the micropub text
| |
| − | ** Tim will send a reviewer suggestion
| |
| − | ** We will not request images mandatory for each micropub
| |
| − | | |
| − | ===GAL4 driver lines===
| |
| − | * ready to submit?
| |
| − | ** will give Paul a deadline for submission
| |
| − | | |
| − | ===Data not shown===
| |
| − | * ok to start contacting publishers to ask their policies?
| |
| − | ** Yes, we should start asking Elsevier and ping again GSA
| |
| − | | |
| − | =March 27 2017=
| |
| − | ==Agenda and Minutes==
| |
| − | | |
| − | ===Minipubs Discussion===
| |
| − | * need to revise the workflow for minipubs before proceeding with Tim's pcn-1 submission
| |
| − | **power point slides here: https://docs.google.com/presentation/d/1GA_9nNtb4OgIP9I64arqHS1jQF2CAqPORY3mogAFz-g/edit#slide=id.p3 (also e-mailed a pdf version of it)
| |
| − | *** We will continue with Tim's submission but we will prioritize focusing on micropubs instead of minis
| |
| − | *** We are not yet ready to handle all the possible data types user might want to submit with minipubs
| |
| − | *** Minipubs should be our final vision and will be able to be assembled when all the micro datasets and submission forms will be in place
| |
| − | | |
| − | ===Sustainability===
| |
| − | * We propose to charge companies an Article Processing Charges (APC) but we will waive the fee for academics.
| |
| − | * Sustainability will be an issue around year 4. If the system will be adopted by the community and we will get enough submissions, wouldn't a 50$ charge/submission be acceptable- also for academia? ~100 submissions/month = 1FTE?
| |
| − | | |
| − | ===Minipublication name===
| |
| − | * If we are not limiting the number of micros in a mini, shall we consider to use a different name for the longer narratives?
| |
| − | * The word 'Mini' alludes to something small or of short length
| |
| − | | |
| − | =March 21 2017=
| |
| − | | |
| − | ==Agenda==
| |
| − | | |
| − | ===Data not shown original Article===
| |
| − | * Tim: Shall we not emphasize in the micropub that the "data not shown" was mentioned in another paper? Will journals think that "data not shown" belongs to that journal?
| |
| − | * The reviewer to which we sent out the cyb-3 micropub last week suggested the opposite: make sure that in the micropub is emphasized that it was data mentioned but not shown in a previous publication. Cite the publication.
| |
| − | * Ask Ruth Isaacson's advice?
| |
| − | | |
| − | ===Micropubs embedded in Minipubs===
| |
| − | * send out for review just the mini pub or each individual micropub first? We touched on the topic last friday (Tim, Karen, Daniela). Tim was suggesting to send out just the mini pub.
| |
| − | * Pros:
| |
| − | ** We use less reviewers
| |
| − | * Cons:
| |
| − | **we normally assign DOIs to the micropubs AFTER the reviewer approves the micropub
| |
| − | ** we think of building up MINIpubs from existing micropubs. In the case we are dealing with, we are defragmenting a longer piece into smaller pieces
| |
| − | ** to mirror the process on how we would deal it in the future, we should send individual findings to reviewers first, we could send a couple of micros to each reviewer: e.g. one will evaluate the 2 expression results, the other will evaluate the phenotype ones.
| |
| − | | |
| − | ===Biocuration poster===
| |
| − | * Will send to Raymond for printing @caltech
| |
| − | * files for future reference here: https://drive.google.com/drive/u/1/folders/0B_LC59YpkJJBUlgxY2Z3S0VybFU
| |
| − | * note that the ppt looks funny in google slides, the pdf version of it is the final look
| |
| − | | |
| − | ===Submissions===
| |
| − | * Received one submission out of 8 requests.
| |
| − | | |
| − | ==Minutes==
| |
| − | ===Data not shown original Article===
| |
| − | * Forget about Data not shown for now and focus on unpublished results (journals may think data not shown belongs to them)
| |
| − | ===Micropubs embedded in Minipubs===
| |
| − | * Send minipubs data just to one reviewer, assign DOIs to each individual finding and to the mini
| |
| − | ===Booth===
| |
| − | * at the booth have an example on how micro and minipubs will look on CVs
| |
| − | | |
| − | =March 8 2017=
| |
| − | | |
| − | ==Agenda and minutes==
| |
| − | * Celebrate the score
| |
| − | ** Paul e-mailed the program director and see what the percentage is
| |
| − | ** critique was good, few minor points to address, e.g.:
| |
| − | *** high throughput data
| |
| − | *** collaboration with publishers -we could contact e-life, collaboration with the Coko foundation deals with the publishing cycle already
| |
| − | *** charging for submissions? yes for companies, not for academia, will say we will waive submission charges for academia
| |
| − | * Booth at the IWM
| |
| − | ** Shall we have one?
| |
| − | *** yes, we will have one, pay through WB
| |
| − | * Booklet at the IWM ([https://github.com/WormBase/micropublications/issues/52 see github #52])
| |
| − | ** Gather micros to generate the booklet
| |
| − | ***booklet: have one of two each flavor submissions
| |
| − | <pre>
| |
| − | data not shown: 1 or more
| |
| − | reagent: e.g. gal-4 drivers
| |
| − | new interesting unpublished data
| |
| − | negative data
| |
| − | ask people what would you like to see published-what you know as common knowledge and has never been published
| |
| − | </pre>
| |
| − | | |
| − | ** Distribute booklet at posters that feature the micros and at booth (WB, Nemametrix, TransgeneOme(?) etc)
| |
| − | * Tim's micropub -how to deal with longer narratives
| |
| − | ** Daniela/Karen will work with Tim to get the atomized data from his submission and will work to generate the MINI-Publication (composed of 2+ micros)
| |
| − | ** Confidence rating discussion (shall we allow authors to put in the confidence level of a result)
| |
| − | * Mitani alleles ([https://github.com/WormBase/micropublications/issues/51 see github #51])
| |
| − | ** Can Paul reach out to Shohei and initiate the collaboration?
| |
| − | ***yes he will. We might set up a conf call with Mitani to discuss it in greater detail
| |
| − | ***we can think of having robot generated descriptions for the mitani alleles
| |
| − | ***if a mitani allele is tested by someone else and gets different results it will be a new micro that will cite the original submission from Mitani
| |
| − | * Alternatives for the current Micropublication: biology site ([https://github.com/WormBase/micropublications/issues/25 see github #25])
| |
| − | **very time consuming right now loading articles into our current site, we need to figure out how to make this more efficient- can we get something better for the 20-50 articles in time for the IWM?
| |
| − | *** Daniel W could dedicate some time on the project at the beginning (manual entry), we will continue to look into alternatives and, if the grant goes in, we will implement this in phase 1
| |
| − | * Other points
| |
| − | ** microreviews
| |
| − | | |
| − | = 2106 meetings=
| |
| − | [[2016]]
| |