Difference between revisions of "Administration:Installing WormMine"
| (111 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | ''This document describes how to install and configure the Intermine instance at WormBase. | |
= Quick Overview = | = Quick Overview = | ||
| + | |||
| + | <span style="color: red">'''Work towards staging and production should be carried out as "intermine" user (<code>sudo su - intermine</code>).'''</span> | ||
| + | |||
| + | <span style="color: red">'''Previous build examples are available at: http://wiki.wormbase.org/index.php/Website:WormMine_Builds'''</span> | ||
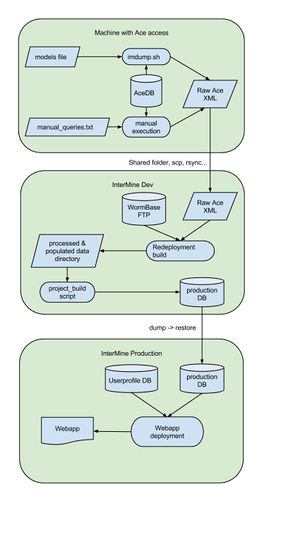
[[File:InterMine workflow.jpg|thumb|Data flow diagram]] | [[File:InterMine workflow.jpg|thumb|Data flow diagram]] | ||
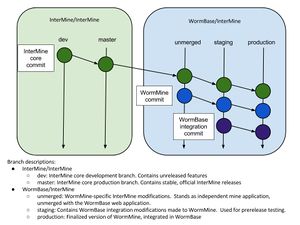
| + | [[File:InterMine branch diagram.jpg|thumb|Branch diagram]] | ||
* InterMine machines: | * InterMine machines: | ||
| − | ** production: 206.108.125.166 | + | ** production: 206.108.125.166 (hostname: wb-intermine) |
| − | ** development: 206.108.125.174 | + | ** development: 206.108.125.174 (hostname: wb-intermine-prod) |
* Important directories: | * Important directories: | ||
** database backup: /nfs/wormbase/wormmine/database_dumps | ** database backup: /nfs/wormbase/wormmine/database_dumps | ||
| Line 44: | Line 49: | ||
= Installation / configuration = | = Installation / configuration = | ||
| + | == Environment Setup == | ||
| + | |||
| + | Add "intermine" user: | ||
| + | |||
| + | <pre> | ||
| + | sudo adduser intermine | ||
| + | </pre> | ||
| + | |||
| + | Password available from Joachim (later: Abigail, Todd). | ||
| + | |||
== Dependencies == | == Dependencies == | ||
=== Git === | === Git === | ||
| Line 58: | Line 73: | ||
Download [http://java.com/en/download/index.jsp here]. | Download [http://java.com/en/download/index.jsp here]. | ||
Since InterMine can be memory intensive, it's helpful to pass environment variables to ant through the <code>ANT_OPTS</code> variable. | Since InterMine can be memory intensive, it's helpful to pass environment variables to ant through the <code>ANT_OPTS</code> variable. | ||
| + | |||
| + | <span style="color: red">'''TODO: figure out the real reason why these parameters should be used. They have little to do with memory usage, but appear to optimize for throughput.'''</span> | ||
| + | |||
| + | [https://intermine.readthedocs.org/en/1.1/system-requirements/software/java/#index-0 Source] | ||
<code>$ export ANT_OPTS="-server -XX:MaxPermSize=256M -Xmx1700m -XX:+UseParallelGC | <code>$ export ANT_OPTS="-server -XX:MaxPermSize=256M -Xmx1700m -XX:+UseParallelGC | ||
-Xms1700m -XX:SoftRefLRUPolicyMSPerMB=1 -XX:MaxHeapFreeRatio=99" | -Xms1700m -XX:SoftRefLRUPolicyMSPerMB=1 -XX:MaxHeapFreeRatio=99" | ||
</code> | </code> | ||
| + | |||
| + | |||
| + | Notes: | ||
| + | You should change the <code>-Xmx</code> and <code>-Xms</code> values if you have very little or very much RAM in your computer. | ||
| + | |||
| + | Increase the <code>MaxPermSize</code> setting if you get this error <code>java.lang.OutOfMemoryError: PermGen space</code> | ||
=== Ant === | === Ant === | ||
| Line 67: | Line 92: | ||
=== Tomcat === | === Tomcat === | ||
| − | |||
| − | + | <pre> | |
| + | # Assuming you cloned website-intermine first: | ||
| + | # git clone https://github.com/WormBase/website-intermine.git | ||
| + | ./website-intermine/scripts/install_tomcat.sh 7.0.47 | ||
| + | </pre> | ||
| + | |||
| + | '''Note:''' the version number (7.0.47) might be out-of-date. To get a listing of currently available versions, type: | ||
<pre> | <pre> | ||
| − | + | ./website-intermine/scripts/install_tomcat.sh | |
| + | </pre> | ||
| + | |||
| + | ==== Starting Tomcat ==== | ||
| + | |||
| + | <pre> | ||
| + | cd apache-tomcat-TOMCATVERSION | ||
| + | ./bin/startup.sh | ||
| + | </pre> | ||
| + | |||
| + | ==== Stopping Tomcat ==== | ||
| + | |||
| + | <pre> | ||
| + | cd apache-tomcat-TOMCATVERSION | ||
| + | ./bin/shutdown.sh | ||
</pre> | </pre> | ||
| Line 78: | Line 122: | ||
<pre> | <pre> | ||
| − | vim apache-tomcat- | + | vim apache-tomcat-TOMCATVERSION/conf/server.xml |
</pre> | </pre> | ||
| Line 94: | Line 138: | ||
For '''user-specific port numbers''' see the Google Doc "Developer Resources at WormBase". | For '''user-specific port numbers''' see the Google Doc "Developer Resources at WormBase". | ||
| − | === | + | === PostgreSQL === |
| + | Refer to [http://intermine.readthedocs.org/en/latest/system-requirements/software/postgres/ InterMine PostgreSQL installation guide] | ||
| − | + | ==== WormMine Workflow ==== | |
| − | |||
| − | |||
| − | |||
| − | ==== | ||
<pre> | <pre> | ||
| − | + | CREATE ROLE intermine_admin WITH SUPERUSER LOGIN PASSWORD 'SECRET'; | |
</pre> | </pre> | ||
| − | |||
| − | |||
| − | |||
=== Perl === | === Perl === | ||
| Line 113: | Line 151: | ||
== Download and Install WormMine == | == Download and Install WormMine == | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<pre> | <pre> | ||
git clone https://github.com/WormBase/website-intermine.git | git clone https://github.com/WormBase/website-intermine.git | ||
| − | + | ./website-intermine/scripts/install_intermine.sh ENVIRONMENT | |
| + | </pre> | ||
| − | + | '''ENVIRONMENT''' is either of the following options: | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | * production | |
| − | + | * staging | |
| − | + | * development | |
| − | |||
=== Create properties file === | === Create properties file === | ||
| Line 178: | Line 174: | ||
** <HELP REQUESTS ARE SENT HERE>: can be same address as above, this is where input from the InterMine help form gets sent. | ** <HELP REQUESTS ARE SENT HERE>: can be same address as above, this is where input from the InterMine help form gets sent. | ||
| − | = | + | = Generate / fetch data files for the build host = |
| − | + | ||
| + | The build requires the following data files. Currently, all of these are retrieved manually from the FTP site except for the Ace XML files. | ||
| − | |||
* Fasta | * Fasta | ||
* GFF3 | * GFF3 | ||
| Line 188: | Line 184: | ||
* Ace XML files | * Ace XML files | ||
| − | + | == 1. Generate ACeDB XML files == | |
| − | + | On a machine that has Acedb installed (such as <code>dev.wormbase.org</code>), check out the [https://github.com/wormbase/website-intermine website-intermine] repository then run: | |
| − | |||
| − | + | <pre> | |
| − | + | $ cd website-intemine | |
| + | $ ./scripts/dump_ace.sh WSXXX | ||
| + | </pre> | ||
| − | + | For the supplied version, this script implements the following steps: | |
| − | + | * Generates generic per-class XML dumps for each species in <code>model</code>. | |
| − | + | * Generates custom XML dumps for specific classes. | |
| − | * | ||
| − | |||
| − | |||
| − | + | XML files will be deposited in /mnt/ephemeral0/intermine-builds/WSXXX | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | For WS246, this step requires ~2 hours and ~18GB of disk space for uncompressed files. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | NOTE FROM TH: | |
| + | dump_ace.sh MAY BE INCOMPLETE. It does not seem to account for all queries in acerb-dev/acedb/manual_queries.txt. What follows is some old but still possibly relevant documentation. We may need to fold remaining queries in manual_queries.txt into dump_ace.sh | ||
| − | + | <pre> | |
| + | The files generated will reflect the ace queries used to generate them. All species are loaded unless otherwise specified. | ||
| + | Each query in website-intermine/acedb-dev/acedb/manual_queries.txt must be run individually in tace, followed by show -x -f <TYPE>.xml where <TYPE> is the ace type being queried. | ||
| + | These queries represent desirable subsets of those represented types. | ||
| + | </pre> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | === QAQC of AceDB XML files === | |
| − | |||
| − | + | There should be one XML file for each class listed in models, plus the special classes handled in dump_ace.sh. Check the start and end of each file to ensure that the dump process for each class completed successfully. | |
| − | |||
| − | + | === Data Delivery === | |
| − | |||
| − | |||
| − | acedb | + | ''Once the acedb data dumps have been generated, they are delivered to the appropriate intermine development instance.'' -- ''THIS SHOULD BE SCRIPTED!'' |
| − | |||
| − | + | <pre> | |
| − | + | rsync -Cav /mnt/ephemeral0/intermine-builds/WSXXX <TARGET MACHINE IP>:/media/ephemeral0/intermine-builds/ | |
| + | </pre> | ||
| − | + | ''Sorry for the path name differences. Will iron this out later.'' | |
| − | |||
| − | + | '''IMPORTANT!''': Please use the INTERNAL IP of the Intermine development instance. Data transfer via internal IPs (as opposed to hostnames or public IPs) is entirely free on AWS. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | TH: DONE THROUGH HERE | |
| − | + | == 2. Acquire and pre-process data files == | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Acquires data files from their appropriate data sources, and pre-processes each one accordingly. | Acquires data files from their appropriate data sources, and pre-processes each one accordingly. | ||
| Line 326: | Line 278: | ||
redeployment]$ '''ant run-all''' | redeployment]$ '''ant run-all''' | ||
| − | Size | + | Size requirements: |
| − | + | ||
| − | + | * WS239: 70G | |
| − | + | * WS240: 75G | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | = Build a new production database = | |
| + | === Custom WormBase loading processes === | ||
| + | ==== Protein Fasta ==== | ||
| + | * custom loader <pre>WormBaseProteinFastaLoaderTask.java</pre> | ||
| + | * puts the first ID of the title row into both Protein.primaryIdentifier and Protein.primaryAccession, and the second ID into protein.secondaryIdentifier | ||
| + | ==== GFF3 ==== | ||
| + | * custom loader <pre>WormbaseGff3CoreGFF3RecordHandler.java</pre> | ||
| + | * when processing transcript record it creates a reference to it's parent gene | ||
| + | * when processing coding sequence record it creates a reference to it's parent transcript | ||
| + | ==== XML ==== | ||
| + | * custom loader <pre>WormbaseAcedbConverter.java</pre> | ||
| + | * can load any XML, not ace specific | ||
| + | * loads intermine class fields as mapped by mapping file | ||
| + | ====Mapping file format:==== | ||
<pre> | <pre> | ||
| − | + | # this is a comment, comment lines are ignored | |
| − | + | # regular annotation, this field will be filled in | |
| − | / | + | # with the value of the evaluated xpath |
| + | primaryIdentifier = /Variation/text()[1] | ||
| + | |||
| + | # returns true if xpath returns any nodes at all, | ||
| + | # useful for data contained in ace tags themselves | ||
| + | if.naturalVariant = /XPATH/... | ||
| − | + | # type casting allowed, this example will add the value as a | |
| + | # Phenotype record | ||
| + | (Phenotype)parents = /XPATH/... | ||
</pre> | </pre> | ||
| + | |||
====== Troubleshooting ====== | ====== Troubleshooting ====== | ||
| Line 367: | Line 336: | ||
=== Build the database === | === Build the database === | ||
| + | |||
| + | <pre> | ||
| + | cd ~/website-intemine | ||
| + | ./scripts/build_intermine.sh WS240 | ||
| + | </pre> | ||
| + | |||
| + | <span style="color: red">'''Quick fix for resolving missing organisms.xml file:'''</span> | ||
| + | |||
| + | <pre> | ||
| + | cd /nfs/wormbase/data/intermine/datadir | ||
| + | mkdir entrez-organism | ||
| + | cp /home/intermine/organisms.xml entrez-organism | ||
| + | </pre> | ||
| + | |||
| + | <span style="color: red">'''The instructions below will be obsolete, once the automatic execution of the script above is confirmed to work.'''</span> | ||
* From the intermine directory, navigate to <code>wormmine</code> | * From the intermine directory, navigate to <code>wormmine</code> | ||
| − | + | ||
| + | <pre> | ||
| + | cd wormmine | ||
| + | mkdir /YOURPATH/datadir/entrez-organism | ||
| + | </pre> | ||
* Run build | * Run build | ||
| − | + | wormmine]$ '''../bio/scripts/project_build -b -v localhost wormmine_wsVERSION_PATCH''' | |
| + | |||
| + | * '''VERSION''': WormBase release version (e.g. WS240) | ||
| + | * '''PATCH''': InterMine build patch, which indicates rebuilds that were requested due to data inconsistencies, data loss, etc. (e.g. 3) | ||
This will run all sources configured in <code>intermine/wormmine/project.xml</code> file. | This will run all sources configured in <code>intermine/wormmine/project.xml</code> file. | ||
| − | + | The project.xml file is described in more detail in the [https://intermine.readthedocs.org/en/1.2.3/get-started/tutorial/#project-xml official documentation]. | |
| + | |||
| + | Issues can be addressed on the InterMine developer list: dev (AT) intermine.org | ||
| + | |||
| + | Output file: <code>wormmine_wsVERSION_PATCH.final</code> | ||
| − | + | Log file: <code>pbuild.log</code> | |
| + | |||
| + | ==== Troubleshooting ==== | ||
| + | |||
| + | If an exceptions occur, then "pbuild.log" should be checked for a more detailed explanation. | ||
==== Build Steps (Preliminary Info) ==== | ==== Build Steps (Preliminary Info) ==== | ||
| − | Building the database refers to the process in which the project build script | + | Building the database refers to the process in which the project build script compiles each source together into a production database. |
| + | |||
| + | <span style="color: red">'''It is unclear which parts of the database are build using "build-db", which cannot recover/be resumed when it fails, and everything coming afterwards.'''</span> | ||
===== 1. Build DB ===== | ===== 1. Build DB ===== | ||
| Line 392: | Line 393: | ||
==== Restarting Building from Milestones/Checkpoints ==== | ==== Restarting Building from Milestones/Checkpoints ==== | ||
| − | If a build error is encountered, it will appear in the output of the build command as "BUILD FAILED". To restart from the latest checkpoint, run the project build script with the -b (build) flag replaced with -l (recover) | + | If a build error is encountered, it will appear in the output of the build command as "BUILD FAILED". To restart from the latest checkpoint, run the project build script with the -b (build) flag replaced with -l (recover). If the -b flag is not omitted, then "build-db" will be run again, which can take a long time. |
<code> | <code> | ||
| Line 412: | Line 413: | ||
== Instantiate database dump == | == Instantiate database dump == | ||
| + | |||
| + | Backup archive can be found at <pre>/nfs/wormbase/wormmine/database_dumps</pre> | ||
| + | |||
To instantiate a previously built WormMine production database. | To instantiate a previously built WormMine production database. | ||
| Line 439: | Line 443: | ||
</pre> | </pre> | ||
| − | == Migrating to production == | + | == Migrating from the unmerged to staging branch == |
| + | |||
| + | git merge is used to move the files and changes on the unmerged branch into staging. | ||
| + | |||
| + | <pre> | ||
| + | [15:32|jdmswong@wb-intermine|webapp]$ git checkout staging | ||
| + | Switched to branch 'staging' | ||
| + | [15:34|jdmswong@wb-intermine|webapp]$ git merge unmerged --squash | ||
| + | ....... | ||
| + | [15:36|jdmswong@wb-intermine|webapp]$ git commit | ||
| + | </pre> | ||
| + | |||
| + | == Migrating to production machine == | ||
The production database must be moved to the production server to be served by the webapp. | The production database must be moved to the production server to be served by the webapp. | ||
| Line 465: | Line 481: | ||
<pre> | <pre> | ||
| − | createdb < | + | createdb -U intermine -E SQL_ASCII wormbase-wsVERSION-PATCH |
| + | pg_restore -U intermine_admin -d wormbase-wsVERSION-PATCH wormbase-wsVERSION-PATCH.final | ||
| + | </pre> | ||
| + | |||
| + | <span style="color: red">'''Note: the psql command below is probably wrong. Will be removed once I can confirm that the pg_restore works.'''</span> | ||
| + | <pre> | ||
psql -U intermine -d <DESTINATION DATABASE NAME> -f <REMOTE DUMPFILE PATH> | psql -U intermine -d <DESTINATION DATABASE NAME> -f <REMOTE DUMPFILE PATH> | ||
</pre> | </pre> | ||
The database is now instantiated on the production machine | The database is now instantiated on the production machine | ||
| + | |||
| + | If the following error is encountered, your postgres user must be granted super user privilege. | ||
| + | |||
| + | <pre> | ||
| + | jbaran@wb-intermine:/nfs/wormbase/archive/jbaran/WS239-2$ psql -U intermine -d joachim-ws239-2 -f wormmine-ws239-2.sql | ||
| + | SET | ||
| + | SET | ||
| + | SET | ||
| + | SET | ||
| + | SET | ||
| + | SET | ||
| + | SET | ||
| + | psql:wormmine-ws239-2.sql:18: ERROR: must be superuser to create a base type | ||
| + | psql:wormmine-ws239-2.sql:27: ERROR: permission denied for language c | ||
| + | ALTER FUNCTION | ||
| + | psql:wormmine-ws239-2.sql:38: ERROR: permission denied for language c | ||
| + | ALTER FUNCTION | ||
| + | psql:wormmine-ws239-2.sql:53: ERROR: must be superuser to create a base type | ||
| + | ALTER TYPE | ||
| + | psql:wormmine-ws239-2.sql:62: ERROR: must be owner of type bioseg | ||
| + | psql:wormmine-ws239-2.sql:71: ERROR: permission denied for language c | ||
| + | ALTER FUNCTION | ||
| + | psql:wormmine-ws239-2.sql:80: ERROR: must be owner of function bioseg_cmp | ||
| + | psql:wormmine-ws239-2.sql:89: ERROR: permission denied for language c | ||
| + | ALTER FUNCTION | ||
| + | psql:wormmine-ws239-2.sql:98: ERROR: must be owner of function bioseg_contained | ||
| + | psql:wormmine-ws239-2.sql:107: ERROR: permission denied for language c | ||
| + | ALTER FUNCTION | ||
| + | psql:wormmine-ws239-2.sql:116: ERROR: must be owner of function bioseg_contains | ||
| + | psql:wormmine-ws239-2.sql:125: ERROR: permission denied for language c | ||
| + | </pre> | ||
=== Configuring mine to database === | === Configuring mine to database === | ||
| − | On production machine, in <pre> | + | On production machine, in <pre>/home/jdmswong/.intermine/wormmine.properties</pre>, set |
<pre>db.production.datasource.databaseName=<DESTINATION DATABASE NAME> </pre> | <pre>db.production.datasource.databaseName=<DESTINATION DATABASE NAME> </pre> | ||
| Line 492: | Line 544: | ||
=== Launch webapp === | === Launch webapp === | ||
| + | * Note: Catalina must be running before you deploy the webapp ([[Administration:Installing_WormMine#About_Catalina |about catalina]]) | ||
| + | ** also, you must [[Administration:Installing_WormMine#Managing_applications |undeploy]] any instances that may currently be running | ||
* Navigate to intermine/wormmine/webapp | * Navigate to intermine/wormmine/webapp | ||
* Launch webapp: | * Launch webapp: | ||
| − | <code> > '''./ | + | <code> > '''./launch_webapp.sh'''</code> |
| + | * Note: for users who aren't JD, this must be run as | ||
| + | <pre> > sudo -ujdmswong ./launch_webapp.sh </pre> | ||
This script contains: | This script contains: | ||
<nowiki> | <nowiki> | ||
| Line 507: | Line 563: | ||
Webapp is standalone. | Webapp is standalone. | ||
| − | === About | + | === About Catalina === |
| − | + | Catalina is Tomcat's servlet container. Catalina implements specs for servlet and JSP. | |
| + | |||
| + | <pre> | ||
| + | $CATALINA_HOME = /home/jdmswong/website-intermine/software/tomcat/apache-tomcat-6.0.36 | ||
| + | </pre> | ||
| + | |||
| + | |||
| + | Catalina must be running before you deploy the webapp | ||
<nowiki> | <nowiki> | ||
$CATALINA_HOME/bin/startup.sh | $CATALINA_HOME/bin/startup.sh | ||
| − | $CATALINA_HOME/bin/shutdown.sh</nowiki> | + | </nowiki> |
| + | |||
| + | |||
| + | In case of problems with deploying the webapp, try restarting Catalina | ||
| + | <nowiki> | ||
| + | $CATALINA_HOME/bin/shutdown.sh | ||
| + | $CATALINA_HOME/bin/startup.sh | ||
| + | </nowiki> | ||
| + | |||
| + | |||
| + | ==== Managing applications ==== | ||
| + | You can use the Tomcat Web Application manager to view/start/stop/undeploy any applications that may be running. | ||
| + | * Production: <code>http://206.108.125.166:8080/manager/html</code> | ||
| + | * The username and password can be found in <code>/home/jdmswong/website-intermine/software/tomcat/apache-tomcat-6.0.36/conf/tomcat-users.xml</code> | ||
| + | ** look for <code>roles="manager-gui"</code> | ||
| + | You will need to <b>undeploy</b> <code>/tools/wormmine</code> every time you restart the web app. You can do this by clicking on 'undeploy' from the list of applications. | ||
| + | |||
| + | === WebApp Logging === | ||
| + | |||
| + | Logs can be found: | ||
| + | <pre> | ||
| + | intermine/wormmine/webapp/intermine.log | ||
| + | <$CATALINA_HOME>/logs | ||
| + | </pre> | ||
| + | |||
| + | Note: | ||
| + | <pre> | ||
| + | $CATALINA_HOME = /home/jdmswong/website-intermine/software/tomcat/apache-tomcat-6.0.36 | ||
| + | </pre> | ||
| + | |||
| + | TODO: document which logs record what | ||
| + | |||
| + | |||
| + | == Restarting postgres == | ||
| + | In case you ever need to restart postgres | ||
| + | <pre> | ||
| + | sudo -upostgres /etc/init.d/postgresql restart | ||
| + | </pre> | ||
= Attach to WormBase instance = | = Attach to WormBase instance = | ||
| Line 535: | Line 635: | ||
== Reconfigure properties file == | == Reconfigure properties file == | ||
| + | Properties file located <code>~/.intermine/wormmine.properties</code> (currently <code>/home/jdmswong/.intermine/wormmine.properties/</code>) | ||
| + | |||
* Needs to deploy at tools/wormmine | * Needs to deploy at tools/wormmine | ||
webapp.path=tools/wormmine | webapp.path=tools/wormmine | ||
| − | * | + | * Update url as appropriate: |
| + | * For deployment on staging.wormbase.org: | ||
<nowiki> | <nowiki> | ||
webapp.baseurl=http://staging.wormbase.org | webapp.baseurl=http://staging.wormbase.org | ||
| − | webapp.returnurl=http://staging.wormbase.org/auth/openid?openid_identifier=https://www.google.com/accounts/o8/id&redirect=http:// | + | webapp.returnurl=http://staging.wormbase.org/auth/openid?openid_identifier=https://www.google.com/accounts/o8/id&redirect=http://staging.wormbase.org/tools/wormmine/mymine.do# |
| + | |||
| + | project.sitePrefix=http://staging.wormbase.org/tools/wormmine | ||
| + | </nowiki> | ||
| + | |||
| + | * For deployment at www.wormbase.org | ||
| + | <nowiki> | ||
| + | webapp.baseurl=http://www.wormbase.org | ||
| + | webapp.returnurl=http://www.wormbase.org/auth/openid?openid_identifier=https://www.google.com/accounts/o8/id&redirect=http://www.wormbase.org/tools/wormmine/mymine.do# | ||
| + | |||
| + | project.sitePrefix=http://www.wormbase.org/tools/wormmine | ||
</nowiki> | </nowiki> | ||
== Modify wormbase.conf == | == Modify wormbase.conf == | ||
To enable login system, make sure config flag: <code> wormmine_path = 'tools/wormmine' </code> is uncommented. | To enable login system, make sure config flag: <code> wormmine_path = 'tools/wormmine' </code> is uncommented. | ||
| + | |||
| + | = Upgrading between releases = | ||
| + | |||
| + | This process has not been fully automated yet, and thus requires some manual work. | ||
| + | |||
| + | == Upgrade release database == | ||
| + | Instructions above | ||
| + | |||
| + | == Upgrade wormmine.properties file == | ||
| + | The current release uses /home/jdmswong/.intermine/wormmine.properties. | ||
| + | |||
| + | Relevant properties: | ||
| + | <pre> | ||
| + | # this is the production database the mine will use | ||
| + | db.production.datasource.databaseName=wormmine-ws238-3 | ||
| + | |||
| + | # This appears at the top next to Version WS | ||
| + | project.releaseVersion= 238 IM v1.2.1 | ||
| + | </pre> | ||
| + | |||
| + | == Update genomic_model.xml == | ||
| + | This is the central model file, used by the webapp process. It must be imported from the machine which produced the production database. | ||
| + | |||
| + | Note: it can be added to the repository, but updates after each test build. This led to extraneous commits and merging conflicts. | ||
| + | |||
| + | In /home/jdmswong/intermine/wormmine/dbmodel/build/model | ||
| + | <pre> | ||
| + | [17:44|jdmswong@wb-intermine|model]$ rsync 206.108.125.174:/home/jdmswong/idev/wormmine/dbmodel/build/model/genomic_model.xml . | ||
| + | </pre> | ||
| + | |||
| + | = Development in a Nutshell = | ||
| + | |||
| + | <span style="color: red">'''Please provide a walkthrough on how new AceDB class could be added to WormMine. List which files need to be added (incl. why), which configuration files need to be updated, and how https://github.com/WormBase/intermine/blob/unmerged/build_config/wormbase-acedb/Gene_mapping.properties mappings work.'''</span> | ||
| + | |||
| + | <span style="color: red">'''Property files are projecting columns (?) to Ace values via XPath expressions. It is unclear where the data ends up though.'''</span> | ||
| + | |||
| + | [[Category: Architecture (Web Dev)]] | ||
Latest revision as of 23:06, 25 December 2014
This document describes how to install and configure the Intermine instance at WormBase.
Contents
- 1 Quick Overview
- 2 Requirements
- 3 Installation / configuration
- 4 Generate / fetch data files for the build host
- 5 Build a new production database
- 6 Attach to WormBase instance
- 7 Upgrading between releases
- 8 Development in a Nutshell
Quick Overview
Work towards staging and production should be carried out as "intermine" user (sudo su - intermine).
Previous build examples are available at: http://wiki.wormbase.org/index.php/Website:WormMine_Builds
- InterMine machines:
- production: 206.108.125.166 (hostname: wb-intermine)
- development: 206.108.125.174 (hostname: wb-intermine-prod)
- Important directories:
- database backup: /nfs/wormbase/wormmine/database_dumps
- in intermine directory:
- datadir: holds data files
- build_config: config files for build
- redeployment: mine update build
Login information is available in a mine's ~/.intermine/wormmine.properties file.
Requirements
Hardware
Linux
- 8 cores
- 24GB RAM
- ~ 1TB storage
Software
Necessary software and versions:
| Software | Minimum Version | Purpose |
|---|---|---|
| Git | 1.7 | check out and update source code |
| Java SDK | 6.0 | build and use InterMine |
| Ant | 1.8 | invokes the InterMine build |
| Tomcat | 6.0.29 | website |
| PostgreSQL | 8.3 | database |
| Perl | 5.8.8 | run build scripts |
Installation / configuration
Environment Setup
Add "intermine" user:
sudo adduser intermine
Password available from Joachim (later: Abigail, Todd).
Dependencies
Git
Install the command line tool:
$ sudo apt-get install git-core
Configure your user and email:
$ git config --global user.name "Name Surname"
$ git config --global user.email "your.email@gmail.com"
Java
Download here.
Since InterMine can be memory intensive, it's helpful to pass environment variables to ant through the ANT_OPTS variable.
TODO: figure out the real reason why these parameters should be used. They have little to do with memory usage, but appear to optimize for throughput.
$ export ANT_OPTS="-server -XX:MaxPermSize=256M -Xmx1700m -XX:+UseParallelGC
-Xms1700m -XX:SoftRefLRUPolicyMSPerMB=1 -XX:MaxHeapFreeRatio=99"
Notes:
You should change the -Xmx and -Xms values if you have very little or very much RAM in your computer.
Increase the MaxPermSize setting if you get this error java.lang.OutOfMemoryError: PermGen space
Ant
Refer to ant's manual for installation instructions.
Tomcat
# Assuming you cloned website-intermine first: # git clone https://github.com/WormBase/website-intermine.git ./website-intermine/scripts/install_tomcat.sh 7.0.47
Note: the version number (7.0.47) might be out-of-date. To get a listing of currently available versions, type:
./website-intermine/scripts/install_tomcat.sh
Starting Tomcat
cd apache-tomcat-TOMCATVERSION ./bin/startup.sh
Stopping Tomcat
cd apache-tomcat-TOMCATVERSION ./bin/shutdown.sh
Configuring an Alternative HTTP Port
vim apache-tomcat-TOMCATVERSION/conf/server.xml
Replace the port number in this context:
<Service name="Catalina">
<Connector port="YOURPORT" protocol="HTTP/1.1"
connectionTimeout="20000"
redirectPort="8443"
URIEncoding="UTF-8" />
...
For user-specific port numbers see the Google Doc "Developer Resources at WormBase".
PostgreSQL
Refer to InterMine PostgreSQL installation guide
WormMine Workflow
CREATE ROLE intermine_admin WITH SUPERUSER LOGIN PASSWORD 'SECRET';
Perl
Refer to InterMine Perl installation guide
Download and Install WormMine
git clone https://github.com/WormBase/website-intermine.git ./website-intermine/scripts/install_intermine.sh ENVIRONMENT
ENVIRONMENT is either of the following options:
- production
- staging
- development
Create properties file
- Create ~/.intermine directory.
- Copy the sample properties file as
~/.intermine/wormmine.properties - Fill in placeholders as follows:
- <POSTGRES USER PASSWORD>: postgres password for intermine user
- <TOMCAT USER PASSWORD>: tomcat password for intermine user
- <SERVER PUBLIC BASE URL>: Base url of your web server, including port. Sample: http://123.456.789.123:8080
- <CREATE WM ADMIN USERNAME/PASSWORD> create the primary admin account
- <EMAIL ADDRESS TO SEND HELP EMAILS FROM>: your server should be configured to send emails from this address. This will send users password reset emails and the like.
- <HELP REQUESTS ARE SENT HERE>: can be same address as above, this is where input from the InterMine help form gets sent.
Generate / fetch data files for the build host
The build requires the following data files. Currently, all of these are retrieved manually from the FTP site except for the Ace XML files.
- Fasta
- GFF3
- GO ontology
- GO association
- Ace XML files
1. Generate ACeDB XML files
On a machine that has Acedb installed (such as dev.wormbase.org), check out the website-intermine repository then run:
$ cd website-intemine $ ./scripts/dump_ace.sh WSXXX
For the supplied version, this script implements the following steps:
- Generates generic per-class XML dumps for each species in
model. - Generates custom XML dumps for specific classes.
XML files will be deposited in /mnt/ephemeral0/intermine-builds/WSXXX
For WS246, this step requires ~2 hours and ~18GB of disk space for uncompressed files.
NOTE FROM TH:
dump_ace.sh MAY BE INCOMPLETE. It does not seem to account for all queries in acerb-dev/acedb/manual_queries.txt. What follows is some old but still possibly relevant documentation. We may need to fold remaining queries in manual_queries.txt into dump_ace.sh
The files generated will reflect the ace queries used to generate them. All species are loaded unless otherwise specified. Each query in website-intermine/acedb-dev/acedb/manual_queries.txt must be run individually in tace, followed by show -x -f <TYPE>.xml where <TYPE> is the ace type being queried. These queries represent desirable subsets of those represented types.
QAQC of AceDB XML files
There should be one XML file for each class listed in models, plus the special classes handled in dump_ace.sh. Check the start and end of each file to ensure that the dump process for each class completed successfully.
Data Delivery
Once the acedb data dumps have been generated, they are delivered to the appropriate intermine development instance. -- THIS SHOULD BE SCRIPTED!
rsync -Cav /mnt/ephemeral0/intermine-builds/WSXXX <TARGET MACHINE IP>:/media/ephemeral0/intermine-builds/
Sorry for the path name differences. Will iron this out later.
IMPORTANT!: Please use the INTERNAL IP of the Intermine development instance. Data transfer via internal IPs (as opposed to hostnames or public IPs) is entirely free on AWS.
TH: DONE THROUGH HERE
2. Acquire and pre-process data files
Acquires data files from their appropriate data sources, and pre-processes each one accordingly.
On the InterMine machine, in the intermine directory:
- Navigate to the redeployment folder.
intermine]$ cd redeployment/
- The
update.propertiesfile should contain these two entries:
release = WS239
ace-xml-dir = /nfs/wormbase/wormmine/acedb_dumps/${release}
The release is used to generate strings, and must match the format of the FTP site.
ace-xml-dir is where the build looks for the ace xml files generated above. This has to be set to <TARGET MACHINE PATH> from above.
The build downloads and processes fasta, gff3, go, gaf files. In addition to copying and processing the Ace XML files.
Build configuration
| Property | function |
|---|---|
| datadir | The data directory for WormMine |
| release | WormBase release version to use for paths and filenames |
| backup-dirname | directory to backup old data directory too. |
| genomic-fasta-species-file | species to download and/or process genomic fasta for |
| protein-fasta-species-file | species to download and/or process protein fasta for |
| gff3-species-file | same, for gff3s |
| ace-classes file | ace classes to copy and/or process |
ant -p will display all invokable tasks, available for individual execution.
- Run the build
This will backup the old data directory into backup-dirname, delete it, then download and process all file types.
redeployment]$ ant
To only download and process:
redeployment]$ ant run-all
Size requirements:
- WS239: 70G
- WS240: 75G
Build a new production database
Custom WormBase loading processes
Protein Fasta
- custom loader
WormBaseProteinFastaLoaderTask.java
- puts the first ID of the title row into both Protein.primaryIdentifier and Protein.primaryAccession, and the second ID into protein.secondaryIdentifier
GFF3
- custom loader
WormbaseGff3CoreGFF3RecordHandler.java
- when processing transcript record it creates a reference to it's parent gene
- when processing coding sequence record it creates a reference to it's parent transcript
XML
- custom loader
WormbaseAcedbConverter.java
- can load any XML, not ace specific
- loads intermine class fields as mapped by mapping file
Mapping file format:
# this is a comment, comment lines are ignored # regular annotation, this field will be filled in # with the value of the evaluated xpath primaryIdentifier = /Variation/text()[1] # returns true if xpath returns any nodes at all, # useful for data contained in ace tags themselves if.naturalVariant = /XPATH/... # type casting allowed, this example will add the value as a # Phenotype record (Phenotype)parents = /XPATH/...
Troubleshooting
Malformed JSON string: output produced by "ant run-all"
website-intermine/acedb-dev/intermine/redeployment$ ant run-all
Buildfile: /home/yourusername/src/intermine/redeployment/build.xml
get-assembly-ids:
[exec] malformed JSON string, neither array, object, number, string or atom, at character offset 8424 (before "],\n "full_name..."JSON retrieved from ftp://ftp.wormbase.org/pub/wormbase/releases/WS239/species/ASSEMBLIES.WS239.json
[exec] ) at gen_assemblies.pl line 19.
BUILD FAILED
/home/yourusername/src/intermine/redeployment/build.xml:39: exec returned: 255
Total time: 2 seconds
Fix: Contact Kevin Howe so that the JSON file on the FTP server can be updated. JSON formatting errors occur sometimes when the file is manually edited after its generation.
Build the database
cd ~/website-intemine ./scripts/build_intermine.sh WS240
Quick fix for resolving missing organisms.xml file:
cd /nfs/wormbase/data/intermine/datadir mkdir entrez-organism cp /home/intermine/organisms.xml entrez-organism
The instructions below will be obsolete, once the automatic execution of the script above is confirmed to work.
- From the intermine directory, navigate to
wormmine
cd wormmine mkdir /YOURPATH/datadir/entrez-organism
- Run build
wormmine]$ ../bio/scripts/project_build -b -v localhost wormmine_wsVERSION_PATCH
- VERSION: WormBase release version (e.g. WS240)
- PATCH: InterMine build patch, which indicates rebuilds that were requested due to data inconsistencies, data loss, etc. (e.g. 3)
This will run all sources configured in intermine/wormmine/project.xml file.
The project.xml file is described in more detail in the official documentation.
Issues can be addressed on the InterMine developer list: dev (AT) intermine.org
Output file: wormmine_wsVERSION_PATCH.final
Log file: pbuild.log
Troubleshooting
If an exceptions occur, then "pbuild.log" should be checked for a more detailed explanation.
Build Steps (Preliminary Info)
Building the database refers to the process in which the project build script compiles each source together into a production database.
It is unclear which parts of the database are build using "build-db", which cannot recover/be resumed when it fails, and everything coming afterwards.
1. Build DB
Invoked by the -b switch to project_build.
Runs: cd dbmodel ; ant clean build-db
This generates the primary mine model file, and creates a fresh database with the desired schema. To generate the model it merged the model additions for all source types used in the main project.xml file. Ace XML, Gff3, and Fasta sources are the most essential.
Restarting Building from Milestones/Checkpoints
If a build error is encountered, it will appear in the output of the build command as "BUILD FAILED". To restart from the latest checkpoint, run the project build script with the -b (build) flag replaced with -l (recover). If the -b flag is not omitted, then "build-db" will be run again, which can take a long time.
../bio/scripts/project_build -v -l localhost wormmine_dump
This will, instead of rebuilding, attempt to restart by reading the last dump database. Dump databases are created by the build script from sources through specifications in the project.xml through the "dump" attribute. An example:
<source name="wb-acedb-Variation" type="wormbase-acedb" dump="true">
properties go here ...
</source>
The builder will create the <DATABASE NAME>:wb-acedb-Variation database once this source is run. If restarting, the builder will find the most recent of these backup databases, clone it, and resume from there.
About the database
The database name is configured in the properties file as: db.production.datasource.databaseName. Each table represents a class in the model, with additional ones representing many-to-many collections, and various metadata. The InterMine development team does not currently advise for developers to modify the backend database due to many layers of inheritance, although questions may be directed to the InterMine developer list at dev (AT) intermine.org.
Instantiate database dump
Backup archive can be found at
/nfs/wormbase/wormmine/database_dumps
To instantiate a previously built WormMine production database.
- Find your favorite release from WORMMINE DB FTP URL (placeholder, no URL exists)
- Create empty DB
> createdb -U intermine -E SQL_ASCII wormmine
- -U: user set to intermine
- -E: character set used
- Unpack and restore DB
> psql -U intermine -d wormmine -f <WORMMINE RELEASE SQL>
- -U: execute as user
- -d: destination DB
- -f: SQL input file
Copy an existing database
Creates a new database with the contents of an already existing database:
createdb -U intermine -W -T EXISTINGDBNAME NEWDBNAME
Note: the existing database needs to be owned by the "intermine" PostgreSQL user. If that is not the case, then it can be set using the following SQL command:
ALTER DATABASE EXISTINGDBNAME OWNER TO intermine
Migrating from the unmerged to staging branch
git merge is used to move the files and changes on the unmerged branch into staging.
[15:32|jdmswong@wb-intermine|webapp]$ git checkout staging Switched to branch 'staging' [15:34|jdmswong@wb-intermine|webapp]$ git merge unmerged --squash ....... [15:36|jdmswong@wb-intermine|webapp]$ git commit
Migrating to production machine
The production database must be moved to the production server to be served by the webapp.
Any paths used in the following commands are arbitrary as long as they are consistent.
Some databases are named along the lines of wormmine-ws239-2. Database names are arbitrary as long as they are referred to correctly in the mine properties file.
Dumping database
On the development machine:
pg_dump -U intermine <DATABASE NAME> -f <PATH TO DUMPFILE>
This created the database dump file.
Transfer this dumpfile to the production machine. Scp is one of the many options: On development machine:
scp <PATH TO DUMPFILE> <REMOTE MACHINE IP>:<REMOTE DUMPFILE PATH>
Restoring database
On production machine:
createdb -U intermine -E SQL_ASCII wormbase-wsVERSION-PATCH pg_restore -U intermine_admin -d wormbase-wsVERSION-PATCH wormbase-wsVERSION-PATCH.final
Note: the psql command below is probably wrong. Will be removed once I can confirm that the pg_restore works.
psql -U intermine -d <DESTINATION DATABASE NAME> -f <REMOTE DUMPFILE PATH>
The database is now instantiated on the production machine
If the following error is encountered, your postgres user must be granted super user privilege.
jbaran@wb-intermine:/nfs/wormbase/archive/jbaran/WS239-2$ psql -U intermine -d joachim-ws239-2 -f wormmine-ws239-2.sql SET SET SET SET SET SET SET psql:wormmine-ws239-2.sql:18: ERROR: must be superuser to create a base type psql:wormmine-ws239-2.sql:27: ERROR: permission denied for language c ALTER FUNCTION psql:wormmine-ws239-2.sql:38: ERROR: permission denied for language c ALTER FUNCTION psql:wormmine-ws239-2.sql:53: ERROR: must be superuser to create a base type ALTER TYPE psql:wormmine-ws239-2.sql:62: ERROR: must be owner of type bioseg psql:wormmine-ws239-2.sql:71: ERROR: permission denied for language c ALTER FUNCTION psql:wormmine-ws239-2.sql:80: ERROR: must be owner of function bioseg_cmp psql:wormmine-ws239-2.sql:89: ERROR: permission denied for language c ALTER FUNCTION psql:wormmine-ws239-2.sql:98: ERROR: must be owner of function bioseg_contained psql:wormmine-ws239-2.sql:107: ERROR: permission denied for language c ALTER FUNCTION psql:wormmine-ws239-2.sql:116: ERROR: must be owner of function bioseg_contains psql:wormmine-ws239-2.sql:125: ERROR: permission denied for language c
Configuring mine to database
On production machine, in
/home/jdmswong/.intermine/wormmine.properties
, set
db.production.datasource.databaseName=<DESTINATION DATABASE NAME>
Create userprofile database
InterMine needs a separate database to track users and their information.
This can be skipped if an existing userprofile database is present.
- Create empty DB
> createdb -U intermine -E SQL_ASCII userprofile-wormmine
- Build the userprofile DB
> cd wormmine/webapp > ant build-db-userprofile
This formats the empty userprofile database for mine use.
About the userprofile database
The database name is set in the properties file as db.userprofile-production.datasource.databaseName. User information is stored in the userprofile table. Tables that begin with "saved" map users to any data they have saved; such as lists, queries, templates, and so on. List data mapping is stores in bagvalues
Launch webapp
- Note: Catalina must be running before you deploy the webapp (about catalina)
- also, you must undeploy any instances that may currently be running
- Navigate to intermine/wormmine/webapp
- Launch webapp:
> ./launch_webapp.sh
- Note: for users who aren't JD, this must be run as
> sudo -ujdmswong ./launch_webapp.sh
This script contains:
ant clean ant -v default remove-webapp release-webapp
Which may be run in sequence instead. These commands clear previous webapp files, remove any existing webapps which may be launched, and compile and release a new webapp.
Test Webapp
You should be able to reach your new instance through <baseurl>/wormmine Webapp is standalone.
About Catalina
Catalina is Tomcat's servlet container. Catalina implements specs for servlet and JSP.
$CATALINA_HOME = /home/jdmswong/website-intermine/software/tomcat/apache-tomcat-6.0.36
Catalina must be running before you deploy the webapp
$CATALINA_HOME/bin/startup.sh
In case of problems with deploying the webapp, try restarting Catalina
$CATALINA_HOME/bin/shutdown.sh $CATALINA_HOME/bin/startup.sh
Managing applications
You can use the Tomcat Web Application manager to view/start/stop/undeploy any applications that may be running.
- Production:
http://206.108.125.166:8080/manager/html - The username and password can be found in
/home/jdmswong/website-intermine/software/tomcat/apache-tomcat-6.0.36/conf/tomcat-users.xml- look for
roles="manager-gui"
- look for
You will need to undeploy /tools/wormmine every time you restart the web app. You can do this by clicking on 'undeploy' from the list of applications.
WebApp Logging
Logs can be found:
intermine/wormmine/webapp/intermine.log <$CATALINA_HOME>/logs
Note:
$CATALINA_HOME = /home/jdmswong/website-intermine/software/tomcat/apache-tomcat-6.0.36
TODO: document which logs record what
Restarting postgres
In case you ever need to restart postgres
sudo -upostgres /etc/init.d/postgresql restart
Attach to WormBase instance
If you want to enable integration with WormBase, follow these steps:
Checkout merged branch
> git checkout remotes/origin/staging Note: checking out 'remotes/origin/staging'. You are in 'detached HEAD' state. You can look around, make experimental changes and commit them, and you can discard any commits you make in this state without impacting any branches by performing another checkout. If you want to create a new branch to retain commits you create, you may do so (now or later) by using -b with the checkout command again. Example: git checkout -b new_branch_name HEAD is now at 611b791... new tests, shell script to run tests > git checkout -b staging Switched to a new branch 'staging'
Reconfigure properties file
Properties file located ~/.intermine/wormmine.properties (currently /home/jdmswong/.intermine/wormmine.properties/)
- Needs to deploy at tools/wormmine
webapp.path=tools/wormmine
- Update url as appropriate:
- For deployment on staging.wormbase.org:
webapp.baseurl=http://staging.wormbase.org webapp.returnurl=http://staging.wormbase.org/auth/openid?openid_identifier=https://www.google.com/accounts/o8/id&redirect=http://staging.wormbase.org/tools/wormmine/mymine.do# project.sitePrefix=http://staging.wormbase.org/tools/wormmine
- For deployment at www.wormbase.org
webapp.baseurl=http://www.wormbase.org webapp.returnurl=http://www.wormbase.org/auth/openid?openid_identifier=https://www.google.com/accounts/o8/id&redirect=http://www.wormbase.org/tools/wormmine/mymine.do# project.sitePrefix=http://www.wormbase.org/tools/wormmine
Modify wormbase.conf
To enable login system, make sure config flag: wormmine_path = 'tools/wormmine' is uncommented.
Upgrading between releases
This process has not been fully automated yet, and thus requires some manual work.
Upgrade release database
Instructions above
Upgrade wormmine.properties file
The current release uses /home/jdmswong/.intermine/wormmine.properties.
Relevant properties:
# this is the production database the mine will use db.production.datasource.databaseName=wormmine-ws238-3 # This appears at the top next to Version WS project.releaseVersion= 238 IM v1.2.1
Update genomic_model.xml
This is the central model file, used by the webapp process. It must be imported from the machine which produced the production database.
Note: it can be added to the repository, but updates after each test build. This led to extraneous commits and merging conflicts.
In /home/jdmswong/intermine/wormmine/dbmodel/build/model
[17:44|jdmswong@wb-intermine|model]$ rsync 206.108.125.174:/home/jdmswong/idev/wormmine/dbmodel/build/model/genomic_model.xml .
Development in a Nutshell
Please provide a walkthrough on how new AceDB class could be added to WormMine. List which files need to be added (incl. why), which configuration files need to be updated, and how https://github.com/WormBase/intermine/blob/unmerged/build_config/wormbase-acedb/Gene_mapping.properties mappings work.
Property files are projecting columns (?) to Ace values via XPath expressions. It is unclear where the data ends up though.