WormMine
Contents
Current status
|
ONLINE |
Links
wormbase-acedb model additions
Reaching our beta release
Issues for curators
Self-assignment
- Publish lists & templates
Data contained in WormMine
This lists all data sources contained in WormMine
| Source | Description | Source | Species | Data mapping |
|---|---|---|---|---|
| GO | Ontology terms and relationships comprising GO | GO project website. | species neutral | |
| GO Annotations | Relationships between Genes and GO | GO project website. | Caenorhabditis elegans | |
| Genomic sequences | Fasta DNA sequences | WormBase FTP | Caenorhabditis elegans | |
| Protein sequences | Fasta peptide sequences | WormBase FTP | Caenorhabditis elegans | |
| Gene locations | Gene chromosomal coordinates | WormBase FTP GFF3 | Caenorhabditis elegans | |
| Transcript locations | Transcript chromosomal coordinates | WormBase FTP GFF3 | Caenorhabditis elegans | |
| CDS locations | CDS chromosomal coordinates | WormBase FTP GFF3 | Caenorhabditis elegans | |
| Gene metadata | Select fields extracted from Ace | AceDB XML dump | All in AceDB | LINK |
| Transcript metadata | Select fields extracted from Ace | AceDB XML dump | All in AceDB | LINK |
| CDS metadata | Select fields extracted from Ace | AceDB XML dump | All in AceDB | LINK |
| Variation metadata | Select fields extracted from Ace | AceDB XML dump | All in AceDB | LINK |
| Protein metadata | Select fields extracted from Ace | AceDB XML dump | All in AceDB | LINK |
| Phenotype metadata | Select fields extracted from Ace | AceDB XML dump | All in AceDB | LINK |
| Expression Pattern metadata | Select fields extracted from Ace | AceDB XML dump | All in AceDB | LINK |
| Anatomy Term metadata | Select fields extracted from Ace | AceDB XML dump | All in AceDB | LINK |
| Expression Cluster metadata | Select fields extracted from Ace | AceDB XML dump | All in AceDB | LINK |
| Life Stage metadata | Select fields extracted from Ace | AceDB XML dump | All in AceDB | LINK |
| Species data | Name and Taxon ID | AceDB XML dump | All in AceDB | LINK |
What is this data mapping?
A loading program plugin has been created for InterMine which extracts data embedded in XML files directly into an InterMine instance. Mapping files are used to configure this program and detail the AceDB XML dumps to InterMine translation. XPath is used to query the XML, and can be reviewed here.
Understanding our model
The data contained in WormMine follows a central model schema. This model should be understood sufficiently to be able to query the data and create templates.
This schema file contains all of the data types contained in WormMine, relationships between them, and each one's data fields.
How to read it
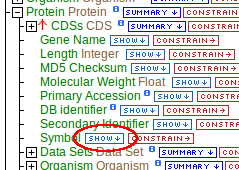
Looking at the protein class:
<class name="Protein" extends="BioEntity" is-interface="true">
<attribute name="molecularWeight" type="java.lang.Float"/>
<attribute name="md5checksum" type="java.lang.String"/>
<attribute name="length" type="java.lang.Integer"/>
<attribute name="geneName" type="java.lang.String"/>
<attribute name="primaryAccession" type="java.lang.String"/>
<reference name="sequence" referenced-type="Sequence"/>
<collection name="CDSs" referenced-type="CDS" reverse-reference="protein"/>
<collection name="genes" referenced-type="Gene" reverse-reference="proteins"/>
<collection name="transcripts" referenced-type="Transcript" reverse-reference="protein"/>
</class>
Line by line:
<class name="Protein" extends="BioEntity" is-interface="true">
extends="BioEntity": Protein is a child of BioEntity therefore it inherits BioEntity's data fields.
Protein's parent, BioEntity:
<class name="BioEntity" is-interface="true">
<attribute name="secondaryIdentifier" type="java.lang.String"/>
<attribute name="symbol" type="java.lang.String"/>
<attribute name="primaryIdentifier" type="java.lang.String"/>
<attribute name="lastUpdated" type="java.util.Date"/>
<attribute name="name" type="java.lang.String"/>
<reference name="organism" referenced-type="Organism"/>
<collection name="synonyms" referenced-type="Synonym" reverse-reference="subject"/>
<collection name="publications" referenced-type="Publication" reverse-reference="bioEntities"/>
<collection name="ontologyAnnotations" referenced-type="OntologyAnnotation" reverse-reference="subject"/>
<collection name="phenotypesObserved" referenced-type="Phenotype" reverse-reference="observedIn"/>
<collection name="phenotypesNotObserved" referenced-type="Phenotype" reverse-reference="notObservedIn"/>
<collection name="crossReferences" referenced-type="CrossReference" reverse-reference="subject"/>
<collection name="dataSets" referenced-type="DataSet" reverse-reference="bioEntities"/>
<collection name="locatedFeatures" referenced-type="Location" reverse-reference="locatedOn"/>
<collection name="locations" referenced-type="Location" reverse-reference="feature"/>
</class>
Protein contains copies of all these attributes, references, and collections for itself. If BioEntity inherits any fields itself, those are included as well.
<attribute name="primaryAccession" type="java.lang.String"/>: This creates an attribute of protein called primaryAccession (read primary accession) which is a string (word(s)). This line enables every protein object to hold a primaryAccession value in addition to any children which may inherit from it.
<reference name="sequence" referenced-type="Sequence"/>: This creates a reference named "sequence" to another data type, in this case Sequence. Only one sequence object can be referenced this way at a time. reverse-reference attributes may appear here, which matches the reciprocal relationship in the referenced type if one exists.
<collection name="CDSs" referenced-type="CDS" reverse-reference="protein"/>: CDSs collection. It can hold many references to CDSs, which in return are stored in the CDS "protein" (CDS.protein) field.
Using QueryBuilder
Use case: How do I find all proteins for a gene?
How are they intuitively connected? Genes are transcribed into transcripts which contain coding sequences which are translated into proteins. Keep this in mind while constructing a query.
- Select "Gene" in the box under "Select a Data Type to Begin a Query"
- Click "Select", or double click "Gene"
This list represents the Gene data available to display. Various fields can be chosen to be shown in the results table, links to other data types followed, and filters can be set to constrain the results.
Desired results?
We want to display the symbols of each involved object, plus the gene ID. This calls for:
- Gene.primaryIdentifier
- Gene.symbol
- Gene (some relationship(s))-> Transcript.symbol
- Gene (some relationship(s))-> CDS.symbol
- Gene (some relationship(s))-> Protein.symbol
Building the query
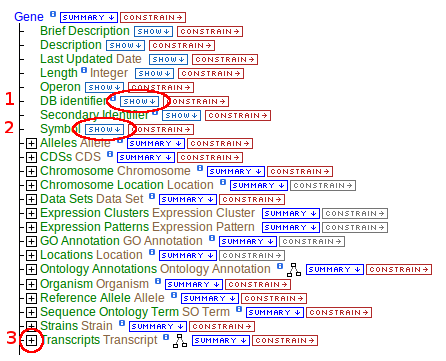
The DB Identifier represents the primaryIdentifier, this is the only field name replaced in this way. Click *SHOW* next to any of these field names to add that attribute to the result table as a column.
- Show the primaryIdentifier (DB Identifier), and symbol
- Gene contains a collection of transcripts, this relationship represents transcription. Expand the relationship by clicking on the
[+]
- Follow these steps for each of the data types in the chain as illustrated.
Running the query at this point will give you results for all genes. If you have a smaller set in mind, like restricting the genes to egl-19 only, a constraint must be set.
- Click "constrain" next to the Gene.symbol field (circle #2), type "egl-19" in the text box, make sure the operator is "=", then "add to query".
Your query overview should resemble this:
- Show results will execute the completed query.
constraining to a list
If you have access to any lists, the constraint dialog box will provide options with respect to them. Fields can be constrained such that their values must or mustn't be a member of that list.
Creating templates
Template queries are predefined (canned) queries. A template query can address a specific question and it can also be a good jump off point for refinements and for constructing queries that answer related questions. To create a template:
* Log in * Construct a query using Query Builder * Must include at least one constrain condition (e.g. restrict the format of the identifier) * "Start building a template query" * Fill in Name, Title and Description, and optionally comment * Make necessary adjustments, then "Save template" * Admin users can make a personal template public by My Mine -> Templates -> Add Tags -> New tag, type in the text box "im:public".
Creating lists
A list is a collection of identifiers of the same type of entities (e.g. genes, proteins, body parts). Lists can be compared and combined and be used as the starting points of queries. Sources of lists may be external (from a third party resource) or from WormMine. To generate a list and make it public:
1. Log in. 2. Make a query (Query Builder). 3. On the Result page, top right corner, select "Create / Add to List" -> "Create New List" -> "All of Columns ...", or "Choose individual items from the table". "Choose individual items from the table" allows further refinement. 4. Provide for the list: a Name, an informative Description, and Tags. Add one tag at a time. Tags help to group lists. For the list to be viewable by the Public, add a tag "im:public". Hit "Create". (This tag should be available for the next template you create). 5. Descriptions and tags can also be edited after a list is saved.
Other Sites
Tutorial on making templates http://www.yeastgenome.org/help/video-tutorials/yeastmine