New 2012 Curation Status
Curation Status & Statistics Form (2012)
The sandbox/testing form can be found here
The CGI code is located on Tazendra/Mangolassi here:
/home/postgres/public_html/cgi-bin/curation_status.cgi
Contents
Pages of the Curation Status Form
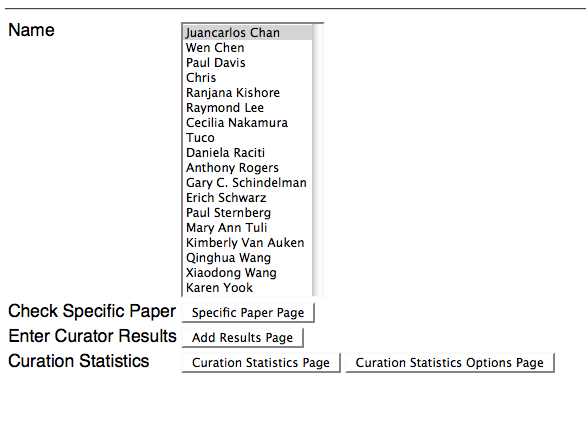
Main Page
Above is a screenshot of the main page of the Curation Status Form. The user/curator is requested to identify who they wish to login as, and to select one of four options to continue:
1) Specific Paper Page - This is where the curator can specify one or more specific papers they wish to view curation status results for (see below).
2) Add Results Page - This is where the curator can add curation status results for one or more specific papers (see below).
3) Curation Statistics Page - This is where the curator can view all curation statistics for ALL datatypes and ALL flagging methods (see below).
4) Curation Statistics Options Page - As an alternative to viewing the curation statistics for ALL datatypes and ALL flagging methods (as with option #3 above), this is where the curator can specify which datatypes and flagging methods they would like to see curation statistics for (see below).
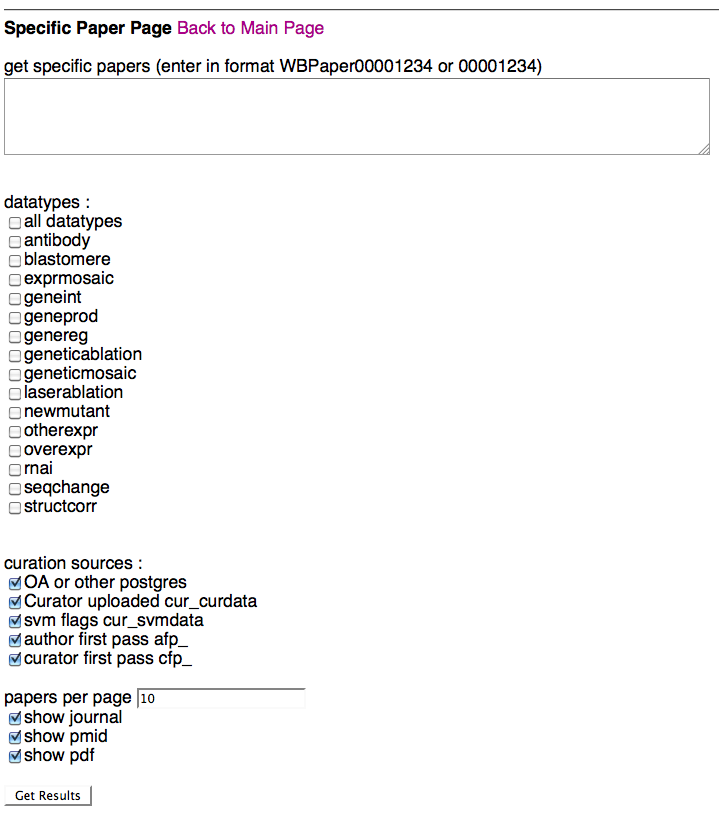
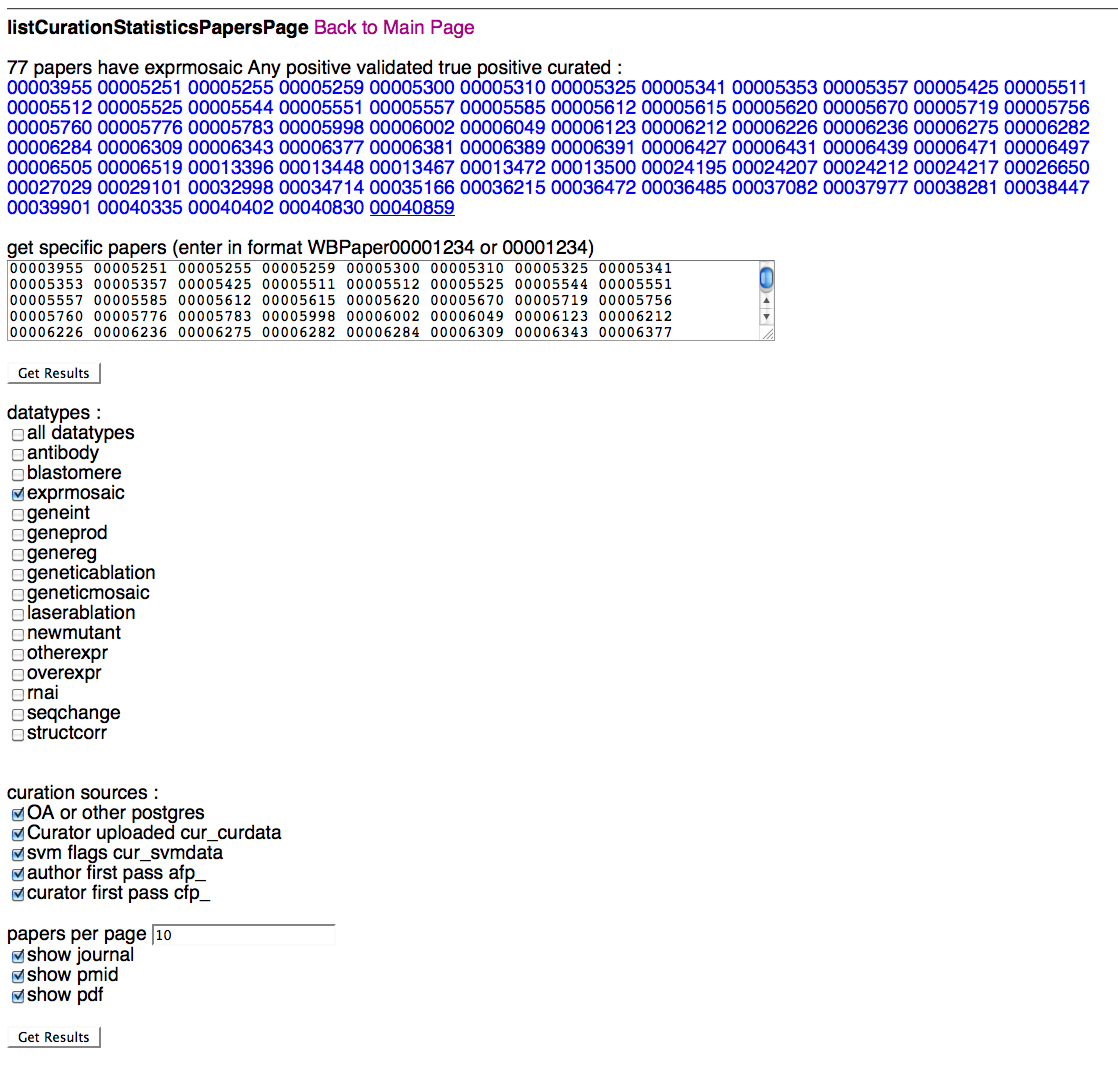
Specific Paper Page
Above is a screenshot of the Specific Paper Page where a curator can specify which paper(s) they would like to view curation status results for. After typing/pasting in one or more WBPaper IDs in the paper entry field, the curator can specify which datatypes and flagging methods they would like to see results for. Note that selecting "all datatypes" will override any single datatype selections below. A curator can select what curation data sources they would like to see results for (i.e. Ontology Annotator and/or cur_curdata), flagging methods (SVM, AFP, CFP), the number of papers they would like to load at one time (default of 10), and whether they would like to see info (and links) for the PubMed ID (PMID), the PDF, and the paper's journal.
Once a curator clicks on "Get Results", they will be directed to the Detailed Results of Papers Page, where they can view the results of their query.
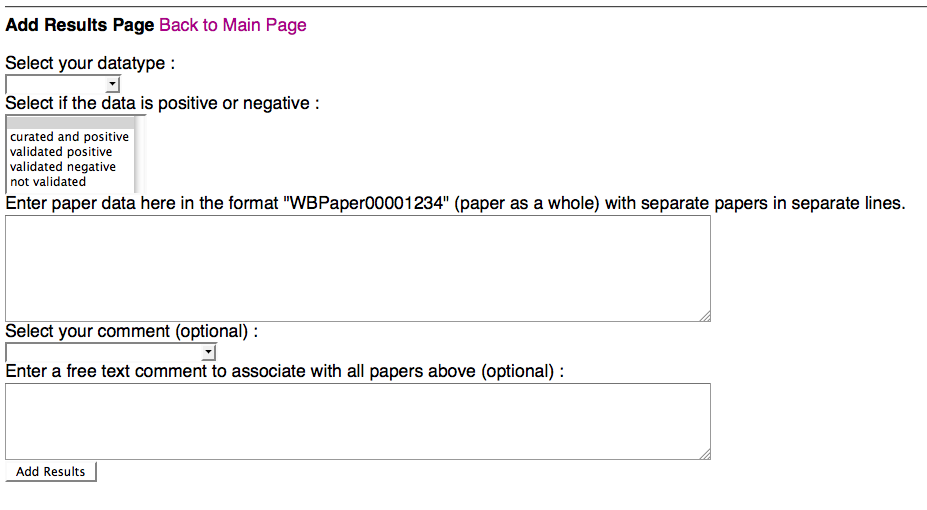
Add Results Page
Above is a screenshot of the Add Results Page of the form, where a curator can add new curation status results for one or more papers that they specify. A curator must specify what datatype they wish to submit paper results for and what the status is for the papers: curated and (hence) positive, validated postive (but not yet curated), validated negative, or (if they need to revert back to not validated, or blank, status) not validated. The curator must then also specify at least one paper for which to apply this curation status in the paper entry field.
Optionally, a curator can select a pre-made comment from a drop down menu and/or enter a free-text comment. Once the curator clicks "Add Results", they will be directed to a data submission summary page:
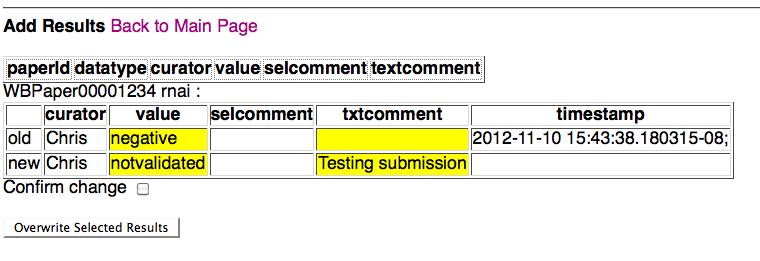
If the results are overwriting existing results, they will be directed to an overwrite confirmation page:
at which point the curator can confirm the overwrite of the previous results for the indicated paper and datatype, or simply go the main page (or go back a page to make corrections/edits). Note that the fields for which data has changed are highlighted in yellow for easy viewing. If the curator confirms the overwrite by checking the confirmation check box and clicking on "Overwrite Selected Results", they will be directed to the overwrite confirmation summary page:
A link is provided to go back to the main page of the form.
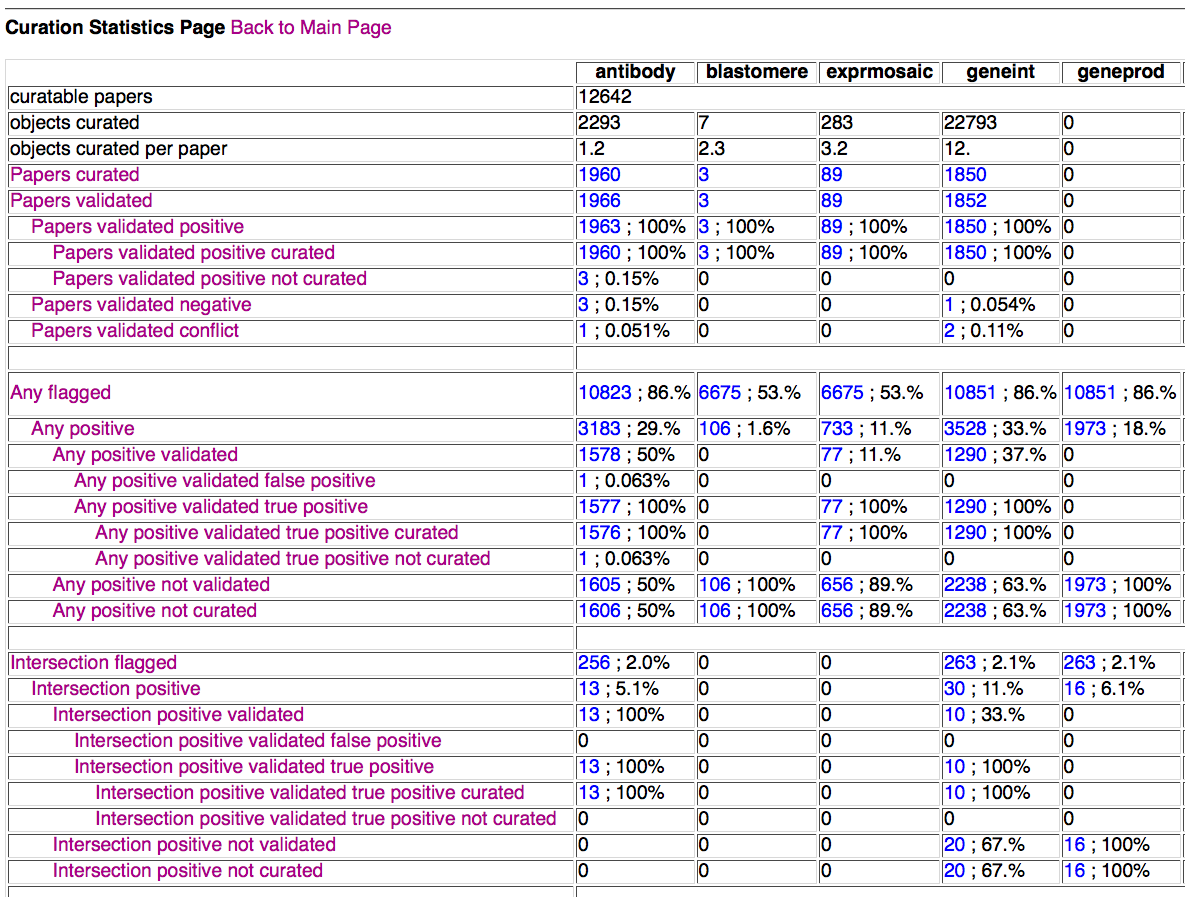
Main Curation Statistics Page
Above is a screenshot of a portion of the entire Curation Statistics table that a curator would be directed to from the main page of the form if they had clicked on the Curation Statistics Page button. Displayed at the top of the table are general paper statistics for a given datatype (datatypes indicated at the top of each column). Below that are statistics for papers that have been flagged (positive or negative) for the indicated datatype by ANY (at least one) flagging method. Below the "Any" statistics are the "Intersection" statistics, indicating papers flagged by ALL flagging methods for the indicated datatype. It should be emphasized here that "flagged" means processed by the flagging method, not necessarily flagged positive. Although not visible in the above screenshot, statistics for SVM results, AFP results, and CFP results are also included in this table.
The "Any", "Intersection", and individual flagging method sections of the table each follow a general template:
Flagged
Flagged Positive
Flagged Positive and Validated
Flagged Positive, Validated False Positive
Flagged Positive, Validated True Positive
Flagged Positive, Validated True Positive, Curated
Flagged Positive, Validated True Positive, Not Curated
Flagged Positive, Not Validated
Flagged Positive, Not Curated
and the individual flagging method sections additionally have a section for flagged negatives:
Flagged Negative
Flagged Negative and Validated
Flagged Negative, Validated True Negative
Flagged Negative, Validated False Negative
Flagged Negative, Validated False Negative, Curated
Flagged Negative, Validated False Negative, Not Curated
Flagged Negative, Not Validated
Flagged Negative, Not Curated
Each row title/header can be clicked on to bring up a small pop-up window with a brief description of what each title means. Each cell of the table has numbers indicating the number of papers that fit the criteria for that datatype and flag status, and the percentage (to two significant digits) that represents of a subset of some larger set. Each percentage is calculated, generally, as follows:
Flagged (% of curatable papers)
Flagged Positive (% flagged)
Flagged Positive and Validated (% flagged positive)
Flagged Positive, Validated False Positive (% flagged positive and validated)
Flagged Positive, Validated True Positive (% flagged positive and validated)
Flagged Positive, Validated True Positive, Curated (% flagged positive and validated true positive)
Flagged Positive, Validated True Positive, Not Curated (% flagged positive and validated true positive)
Flagged Positive, Not Validated (% flagged positive)
Flagged Positive, Not Curated (% flagged positive)
Each cell number (aside from the top three rows) is also a hyperlink to the Prepopulated Specific Papers Page, listing the paper IDs for each paper in the list, as well as providing options for the view of each of those papers in the Detailed Results of Papers Page.
Curation Statistics Page Display Info
The title/headers for each row are displayed at the left AND right sides of the table, to enable easier viewing when there are several datatypes being viewed at once. If the number of datatypes is restricted via the Curation Statistics Options Page, the titles/headers for each row will only display on both left and right sides of the table if more than six datatypes are selected for viewing (this was done to avoid overcrowding of the page when six or fewer datatypes/columns were visible).
The row-title column (leftmost column and, when more than six datatypes are visible, the rightmost column) are set to display at a fixed width of 600 pixels to allow all titles to fit on a single line. All other columns (datatype columns) are set to display at a fixed width of 120 pixels.
Curation Statistics Options Page
Prepopulated Specific Papers Page
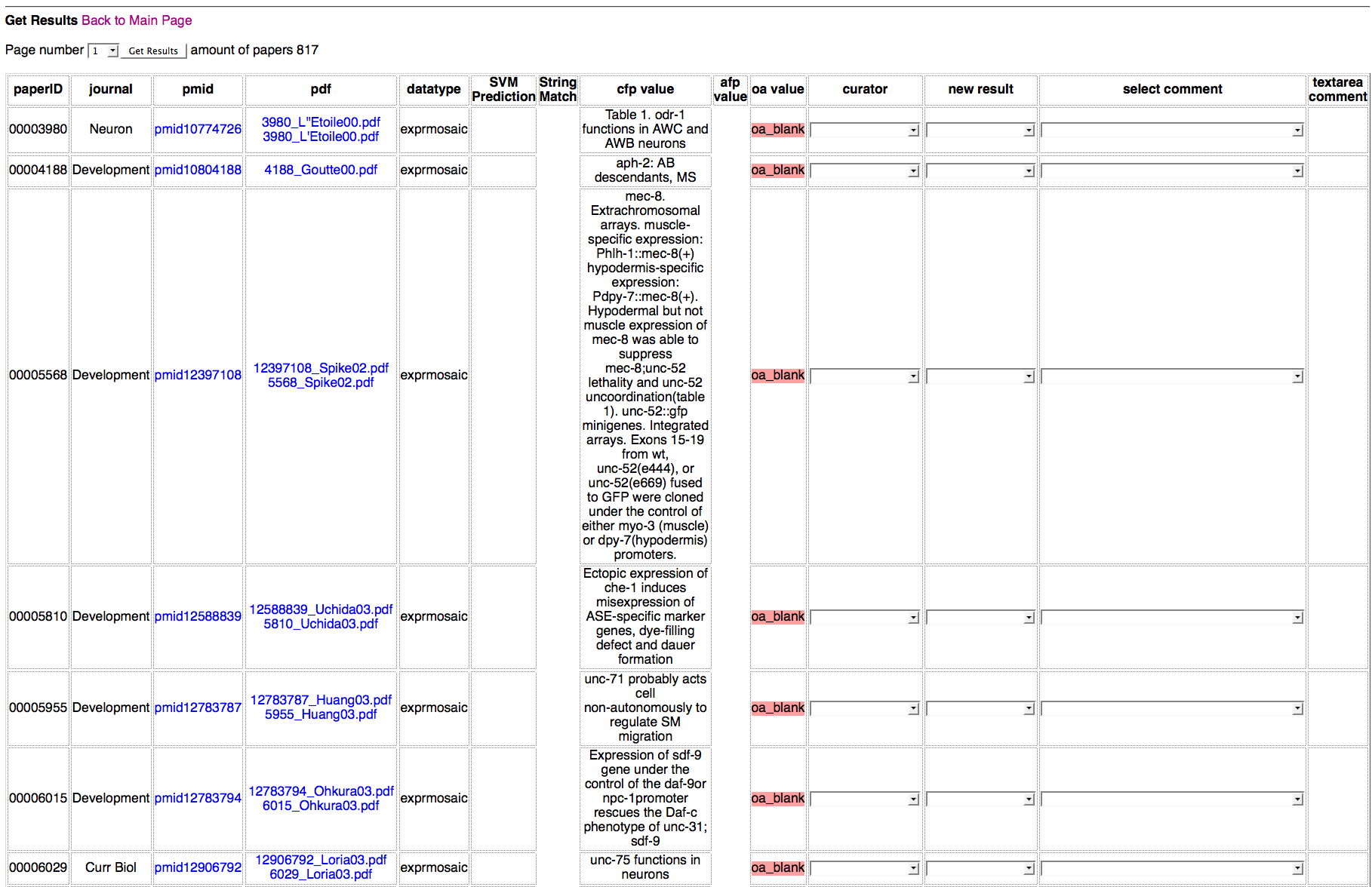
Detailed Results of Papers Page
Code Documentation
Below is the documentation for the form's code, located on Tazendra (when live) or Mangolassi (sandbox):
/home/postgres/public_html/cgi-bin/curation_status.cgi
Precanned Comments
In the Detailed Results of Papers page, curators have the option to select a comment from a drop down list of comments to apply to this paper in the context of the relevant data type.
In the code, the comments are stored in a hash table called %premadeComments. The keys (stored in postgres) of these comments are only numbers, so the descriptions/titles can change or be updated and still apply retroactively.
Code:
sub populatePremadeComments {
$premadeComments{"1"} = "SVM Positive, Curation Negative";
$premadeComments{"2"} = "pre-made comment #2";
$premadeComments{"3"} = "pre-made comment #3";}
So, as of now:
| Key | Comment | | 1 | "SVM Positive, Curation Negative" | | 2 | "pre-made comment #2" | | 3 | "pre-made comment #3" |
Hence, if a completely new comment is desired, a new key will need to be made and there after associated with that new comment. Also, old keys should never be recycled and documentation describing what each key refers to should be maintained in this Wiki.
New Result
Each paper-data-type pair can be assigned a "New Result" indicating its status as curated (or not) or validated (or not), and if validated, positive or negative for the particular paper-data-type pair. These results can be entered via the Add Results Page or directly in the Detailed Results of Papers page via the "New Results" column. The code is below:
Code:
sub populateDonPosNegOptions {
$donPosNegOptions{""} = "";
$donPosNegOptions{"curated"} = "curated and positive";
$donPosNegOptions{"positive"} = "validated positive";
$donPosNegOptions{"negative"} = "validated negative";
$donPosNegOptions{"notvalidated"} = "not validated";}
where "curated", "positive", "negative", and "notvalidated" are the keys (for the %donPosNegOptions hash table in the form code) that will be stored in postgres and the corresponding values (e.g. "curated and positive") are what will be displayed on the form.
Note that "" and "not validated" represent no data for that paper-data-type pair, but "not validated" is present as an option to overwrite accidental validations (it is impossible to go back to a blank "" field via the form).
Data Types
The form determines which data types exist via a 'populateDatatypes' subroutine in the form code. As of 12-5-2012, the form first collects all data types used in SVM from the 'cur_svmdata' postgres table (which, as of 12-5-2012, all also are identically named in the Author First Pass (AFP) and Curator First Pass (CFP) tables) and then supplements with other data types not in SVM but in AFP and CFP (as of 12-5-2012, all anatomy curation related data types) plus one additional data type ("geneticablation") not in SVM, AFP, or CFP.
Here is the code:
sub populateDatatypes {
$result = $dbh->prepare( "SELECT DISTINCT(cur_datatype) FROM cur_svmdata " );
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) { $datatypesAfpCfp{$row[0]} = $row[0]; }
$datatypesAfpCfp{'blastomere'} = 'cellfunc';
$datatypesAfpCfp{'exprmosaic'} = 'siteaction';
$datatypesAfpCfp{'geneticmosaic'} = 'mosaic';
$datatypesAfpCfp{'laserablation'} = 'ablationdata';
foreach my $datatype (keys %datatypesAfpCfp) { $datatypes{$datatype}++; }
$datatypes{'geneticablation'}++;
} # sub populateDatatypes
As for the data types currently (12-5-2012) NOT in SVM but IN AFP and CFP, the data type name is different between the Curation Status form and the AFP and CFP forms. So, the data types named "cellfunc", "siteaction", "mosaic", and "ablationdata" in the AFP and CFP tables are respectively named "blastomere", "exprmosaic", "geneticmosaic", "laserablation" in the Curation Status form.
The IMPORTANT thing here is: if, at some point, the data types are changed (added, renamed, etc.), and the code is not updated in kind, the form will likely break. Curators should tell Juancarlos/Chris/Daniela to update the code.
new datatypes should be accounted in this code :
- - no svm, no afp/cfp : add to %datatypes hash like 'geneticablation'.
- - no svm, yes afp/cfp : add to %datatypesAfpCfp + %datatypes hashes like 'blastomere'
- - yes svm, yes afp/cfp : add to code to populate cur_svmdata, which will populate in the SELECT query
- - yes svm, no afp/cfp : add to code to populate cur_svmdata, which will populate in the SELECT query, but also subsequently delete from %datatypesAfpCfp (to prevent a postgres query to a non-existing table which will crash the form)
Creating PDF links to papers
In the Detailed Results of Papers page, each paper ID is linked to its corresponding PDF document using the code below:
Code:
sub populatePdf {
$result = $dbh->prepare( "SELECT * FROM pap_electronic_path WHERE pap_electronic_path IS NOT NULL");
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
my %temp;
while (my @row = $result->fetchrow) {
my ($data, $isPdf) = &makePdfLinkFromPath($row[1]);
$temp{$row[0]}{$isPdf}{$data}++; }
foreach my $joinkey (sort keys %temp) {
my @pdfs;
foreach my $isPdf (reverse sort keys %{ $temp{$joinkey} }) {
foreach my $pdfLink (sort keys %{ $temp{$joinkey}{$isPdf} }) {
push @pdfs, $pdfLink; } }
my ($pdfs) = join"<br/>", @pdfs;
$pdf{$joinkey} = $pdfs;
} # foreach my $joinkey (sort keys %temp)
} # sub populatePdf
sub makePdfLinkFromPath {
my ($path) = shift;
my ($pdf) = $path =~ m/\/([^\/]*)$/;
my $isPdf = 0; if ($pdf =~ m/\.pdf$/) { $isPdf++; } # kimberly wants .pdf files on top, so need to flag to sort
my $link = 'http://tazendra.caltech.edu/~acedb/daniel/' . $pdf;
my $data = "<a href=\"$link\" target=\"new\">$pdf</a>"; return ($data, $isPdf); }
Note the table name ("pap_electronic_path"), the URL path ("http://tazendra.caltech.edu/~acedb/daniel/"), and (because of the code 'target=\"new\"') that the link will open a new window or tab. Also note that opening another link on the original page (e.g. Detailed Results of Papers page) will open that link in that same new window/tab, clearing out what you had opened previously.
Creating hyperlinks to PubMed paper pages
In the Detailed Results of Papers page each PubMed ID is linked to its corresponding PubMed webpage using the code below:
Code:
sub populatePmid {
$result = $dbh->prepare( "SELECT * FROM pap_identifier WHERE pap_identifier ~ 'pmid'" );
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
my %temp;
while (my @row = $result->fetchrow) { if ($row[0]) {
my ($data) = &makeNcbiLinkFromPmid($row[1]);
$temp{$row[0]}{$data}++; } }
foreach my $joinkey (sort keys %temp) {
my ($pmids) = join"<br/>", keys %{ $temp{$joinkey} };
$pmid{$joinkey} = $pmids;
} # foreach my $joinkey (sort keys %temp)
} # sub populatePmid
sub makeNcbiLinkFromPmid {
my $pmid = shift;
my ($id) = $pmid =~ m/(\d+)/;
my $link = 'http://www.ncbi.nlm.nih.gov/pubmed/' . $id;
my $data = "<a href=\"$link\" target=\"new\">$pmid</a>"; return $data; }
Note the table name ("pap_identifier"), the table specifier ("WHERE pap_identifier ~ 'pmid'"), the URL path ("http://www.ncbi.nlm.nih.gov/pubmed/"), and (because of the code 'target=\"new\"') that the link will open a new window or tab. Also note that opening another link on the original page (e.g. Detailed Results of Papers page) will open that link in that same new window/tab, clearing out what you had opened previously.
Populating the Journal Names
Journal names for each paper are populated via the following code:
sub populateJournal {
$result = $dbh->prepare( "SELECT * FROM pap_journal WHERE pap_journal IS NOT NULL" );
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) { if ($row[0]) { $journal{$row[0]} = $row[1]; } }
} # sub populateJournal
Note the table "pap_journal".
Loading Data into the Form
On the Curation Statistics Options Page, the Specific Paper Page, or the Prepopulated Specific Papers Page, curators have the option to specify what flagging methods (SVM, AFP, and/or CFP), curation sources (Ontology Annotator or cur_curdata [which is the data generated from this form]), and/or data types (e.g. geneint, rnai) they would like to view.
There are separate hashes for storing the different types of data, all of which have a key of datatype, subkey paperID, sub-subkeys of other things depending on the hash (see individual subsections below).
There is an option to select specific datatype, in which case only the data for those datatypes is loaded. Similarly if only some paperIDs have been selected, only those paperIDs are loaded.
Loading curatable papers
Only papers that have a 'valid' pap_status value and a 'primary' pap_primary_data value are considered curatable. These are stored in the %curatablePapers hash. ( paperID => status )
sub populateCuratablePapers {
my $query = "SELECT * FROM pap_status WHERE pap_status = 'valid' AND joinkey IN (SELECT joinkey FROM pap_primary_data WHERE pap_primary_data = 'primary')";
$result = $dbh->prepare( $query );
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) { $curatablePapers{$row[0]} = $row[1]; }
} # sub populateCuratablePapers
Loading afp_ data
Populate %afpEmailed, %afpData, %afpFlagged, %afpPos, %afpNeg.
for each of the chosen datatypes, if they are allowed in %datatypeAfpCfp, query the corresponding afp_ postgres table, and if it's a curatable paper store the value in the %afpData hash (data type, paper ID => AFP result).
Query afp_email and if it's a curatable paper store in %afpEmailed hash ( paperID => 1 ) for afp emailed statistics.
Query afp_lasttouched to see if a paper has been flagged for afp. Skip if it's not a curatable paper. For all %chosenDatatypes store in %afpFlagged ( datatype, paperID => 1 )
For each of the %afpFlagged datatypes that have been chosen (%chosenDatatypes), if there is an %afpData value, store in %afpPos hash ( positive flag for afp ), otherwise store in %afpNeg hash (negative flag for afp ) ( datatype, paperID => 1 )
sub populateAfpData {
foreach my $datatype (sort keys %chosenDatatypes) {
next unless $datatypesAfpCfp{$datatype};
my $pgtable_datatype = $datatypesAfpCfp{$datatype};
$result = $dbh->prepare( "SELECT * FROM afp_$pgtable_datatype" );
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) {
next unless ($curatablePapers{$row[0]});
$afpData{$datatype}{$row[0]} = $row[1]; }
} # foreach my $datatype (sort keys %chosenDatatypes)
$result = $dbh->prepare( "SELECT * FROM afp_email" );
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) {
next unless ($curatablePapers{$row[0]});
$afpEmailed{$row[0]}++; }
$result = $dbh->prepare( "SELECT * FROM afp_lasttouched" );
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) {
next unless ($curatablePapers{$row[0]});
foreach my $datatype (sort keys %chosenDatatypes) {
$afpFlagged{$datatype}{$row[0]}++; } }
foreach my $datatype (sort keys %chosenDatatypes) {
foreach my $joinkey (sort keys %{ $afpFlagged{$datatype} }) {
if ($afpData{$datatype}{$joinkey}) { $afpPos{$datatype}{$joinkey}++; }
else { $afpNeg{$datatype}{$joinkey}++; } } }
} # sub populateAfpData
Loading cfp_ data
Populate %cfpData, %cfpFlagged, %cfpPos, %cfpNeg.
for each of the chosen datatypes, if they are allowed in %datatypeAfpCfp, query the corresponding cfp_ postgres table, and if it's a curatable paper store the value in the %cfpData hash (data type, paper ID => CFP result).
Query cfp_curator to see if a paper has been flagged for cfp. Skip if it's not a curatable paper. For all %chosenDatatypes store in %cfpFlagged ( datatype, paperID => 1 )
For each of the %cfpFlagged datatypes that have been chosen (%chosenDatatypes), if there is an %cfpData value, store in %cfpPos hash ( positive flag for cfp ), otherwise store in %cfpNeg hash (negative flag for cfp ) ( datatype, paperID => 1 )
sub populateCfpData {
foreach my $datatype (sort keys %chosenDatatypes) {
next unless $datatypesAfpCfp{$datatype};
my $pgtable_datatype = $datatypesAfpCfp{$datatype};
$result = $dbh->prepare( "SELECT * FROM cfp_$pgtable_datatype" );
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) {
next unless ($curatablePapers{$row[0]});
$cfpData{$datatype}{$row[0]} = $row[1]; }
} # foreach my $datatype (sort keys %chosenDatatypes)
$result = $dbh->prepare( "SELECT * FROM cfp_curator" );
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) {
next unless ($curatablePapers{$row[0]});
foreach my $datatype (sort keys %chosenDatatypes) {
$cfpFlagged{$datatype}{$row[0]}++; } }
foreach my $datatype (sort keys %chosenDatatypes) {
foreach my $joinkey (sort keys %{ $cfpFlagged{$datatype} }) {
if ($cfpData{$datatype}{$joinkey}) { $cfpPos{$datatype}{$joinkey}++; }
else { $cfpNeg{$datatype}{$joinkey}++; } } }
} # sub populateCfpData
Loading svm data
Populate %svmData hash.
For each of the chosen datatypes, query the cur_svmdata table where cur_datatype is that datatype, and sort by cur_date so that we always have the latest value for a given paper-data-type pair. The svm result is the 4th column, the paper ID is the first column. skip papers that are not %curatablePapers. store in %svmData ( datatype, paper => svm_result ). cur_svmdata could have multiple results for a given paper-data-type pair, we'll consider only the most recent result (by the directory name/date on Yuling's machine).
sub populateSvmData {
# $result = $dbh->prepare( "SELECT * FROM cur_svmdata ORDER BY cur_datatype, cur_date" ); # always doing for all datatypes vs looping for chosen takes 4.66vs 2.74 secs
foreach my $datatype (sort keys %chosenDatatypes) {
$result = $dbh->prepare( "SELECT * FROM cur_svmdata WHERE cur_datatype = '$datatype' ORDER BY cur_date" );
# table stores multiple dates for same paper-datatype in case we want to see multiple results later. if it didn't and we didn't order it would take 2.05 vs 2.74 secs, so not worth changing the way we're storing data
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) {
my $joinkey = $row[0]; my $svmdata = $row[3];
next unless ($curatablePapers{$row[0]});
$svmData{$datatype}{$joinkey} = $svmdata; } }
} # sub populateSvmData
Loading OA data
Populate %objsCurated and %oaData hashes.
Each datatype is stored in different tables and has to be queried separately. The queries are mostly the same.
if ($chosenDatatypes{'newmutant'}) {
$result = $dbh->prepare( "SELECT * FROM app_variation" );
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) { $objsCurated{'newmutant'}{$row[1]}++; }
$result = $dbh->prepare( "SELECT * FROM app_paper" );
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) {
my (@papers) = $row[1] =~ m/WBPaper(\d+)/g;
foreach my $paper (@papers) {
$oaData{'newmutant'}{$paper} = 'curated'; } } }
and similarly for other datatypes.
The example above is for the datatype 'newmutant'. If that datatype is a %chosenDatatypes, query app_variation and store in %objsCurated ( datatype, object => 1 ), then query app_paper matching for WBPaper IDs, and associating to %oaData ( datatype, paperID => 'curated' ).
For other datatypes :
- overexpr : objects from app_transgene ; %oaData from app_paper WHERE joinkey IN (SELECT joinkey FROM app_transgene WHERE app_transgene IS NOT NULL AND app_transgene != ), meaning papers where the postgresID has a corresponding transgene that exists in app_transgene.
- antibody : objects from abp_name ; %oaData from abp_paper
- otherexpr : objects from exp_name ; %oaData from exp_paper
- genereg : objects from grg_name; %oaData from grg_paper
- geneint : objects from int_name; %oaData from int_paper
- rnai : objects from rna_name; %oaData from rna_paper
- blastomere : objects from wbb_wbbtf WHERE joinkey IN (SELECT joinkey FROM wbb_assay WHERE wbb_assay = 'Blastomere_isolation') ; %oaData from wbb_reference WHERE joinkey IN (SELECT joinkey FROM wbb_assay WHERE wbb_assay = 'Blastomere_isolation')
- exprmosaic : objects from wbb_wbbtf WHERE joinkey IN (SELECT joinkey FROM wbb_assay WHERE wbb_assay = 'Expression_mosaic') ; %oaData from wbb_reference WHERE joinkey IN (SELECT joinkey FROM wbb_assay WHERE wbb_assay = 'Expression_mosaic')
- geneticablation : objects from wbb_wbbtf WHERE joinkey IN (SELECT joinkey FROM wbb_assay WHERE wbb_assay = 'Genetic_ablation') ; %oaData from wbb_reference WHERE joinkey IN (SELECT joinkey FROM wbb_assay WHERE wbb_assay = 'Genetic_ablation')
- geneticmosaic : objects from wbb_wbbtf WHERE joinkey IN (SELECT joinkey FROM wbb_assay WHERE wbb_assay = 'Genetic_mosaic') ; %oaData from wbb_reference WHERE joinkey IN (SELECT joinkey FROM wbb_assay WHERE wbb_assay = 'Genetic_mosaic')
- laserablation : objects from wbb_wbbtf WHERE joinkey IN (SELECT joinkey FROM wbb_assay WHERE wbb_assay = 'Laser_ablation') ; %oaData from wbb_reference WHERE joinkey IN (SELECT joinkey FROM wbb_assay WHERE wbb_assay = 'Laser_ablation')
Loading cur_curdata
cur_curdata: this captures all data entered through this form, meaning paper ID, data type, curator ID, validation status (e.g. "curated and positive"), pre-canned comment, and/or free text comment (and timestamp). Note: this table only stores data (and associated paper-data-type pairs) that has been manually entered through this form.
Code:
sub populateCurCurData {
$result = $dbh->prepare( "SELECT * FROM cur_curdata ORDER BY cur_timestamp" ); # in case multiple values get in for a paper-datatype (shouldn't happen), keep the latest
$result->execute() or die "Cannot prepare statement: $DBI::errstr\n";
while (my @row = $result->fetchrow) {
next unless ($chosenPapers{$row[0]} || $chosenPapers{all});
next unless ($chosenDatatypes{$row[1]});
$curData{$row[1]}{$row[0]}{curator} = $row[2];
$curData{$row[1]}{$row[0]}{donposneg} = $row[3];
$curData{$row[1]}{$row[0]}{selcomment} = $row[4];
$curData{$row[1]}{$row[0]}{txtcomment} = $row[5];
$curData{$row[1]}{$row[0]}{timestamp} = $row[6]; }
} # sub populateCurCurData
When populating curator data from curation status, read the cur_curdata postgres table, skip datatypes that were not chosen, skip papers that were not chosen.
Store data in the %curData hash, key is datatype, subkey is paperID, then valuekeys are curator, donposneg (curator result of curated, validatedPos, validatedNeg, notValidated), select comment, text comment, timestamp.
cur_curdata can only have one result for a specific paper-data-type pair, if a new result is entered it will overwrite the previous result.
Processing curated data
The following subroutine will process cur_curdata and oaData into %valCur %valPos %valNeg and into %conflict which has the paper-datatypes that have multiple values, which correspond to a datatype-paper pair's validated+curated, validated+positive, validated+negative.
If a paper has been curated for a datatype, the paper enters into the %valCur AND the %valPos hashes; if it has been validated positive but NOT curated it goes into %valPos ONLY; and if it has been validated negative it will go into %valNeg.
sub populateCuratedPapers {
my ($showTimes, $start, $end, $diff) = (0, '', '', '');
if ($showTimes) { $start = time; }
&populateCurCurData();
if ($showTimes) { $end = time; $diff = $end - $start; $start = time; print "IN populateCuratedPapers populateCurCurData $diff<br>"; }
&populateOa(); # $oaData{datatype}{joinkey} = 'positive';
if ($showTimes) { $end = time; $diff = $end - $start; $start = time; print "IN populateCuratedPapers populateOa $diff<br>"; }
my %allCuratorValues; # $allCuratorValues{datatype}{joinkey} = 0 | 1+
foreach my $datatype (sort keys %oaData) {
foreach my $joinkey (sort keys %{ $oaData{$datatype} }) {
$allCuratorValues{$joinkey}{$datatype}{curated}++; } } # validated positive and curated
foreach my $datatype (sort keys %curData) {
foreach my $joinkey (sort keys %{ $curData{$datatype} }) {
$allCuratorValues{$joinkey}{$datatype}{ $curData{$datatype}{$joinkey}{donposneg} }++; } }
foreach my $joinkey (sort keys %allCuratorValues) {
foreach my $datatype (sort keys %{ $allCuratorValues{$joinkey} }) {
my @values = keys %{ $allCuratorValues{$joinkey}{$datatype} };
if (scalar @values > 1) { $conflict{$datatype}{$joinkey}++; }
else {
my $value = shift @values;
$validated{$datatype}{$joinkey} = $value;
if ($value eq 'curated') { $valPos{$datatype}{$joinkey} = $value; $valCur{$datatype}{$joinkey} = $value; }
elsif ($value eq 'positive') { $valPos{$datatype}{$joinkey} = $value; }
elsif ($value eq 'negative') { $valNeg{$datatype}{$joinkey} = $value; }
} } }
if ($showTimes) { $end = time; $diff = $end - $start; $start = time; print "IN populateCuratedPapers categorizing hash $diff<br>"; }
} # sub populateCuratedPapers
Curation Statistics Calculations
The way that each value is calculated for Curation Statistics table is based on what papers (or, more specifically, paper IDs) populate each of a number of tables. The following hash tables capture validation status:
%valCur - All papers that have been curated for a given datatype %valPos - All papers that have been validated positive for a given datatype, but not yet curated %valNeg - All papers that have been validated negative for a given datatype
When determining, for a particular flagging method, the validation and curation statistics with respect to flagging status, these tables are compared to the table for flagging results to generate the numbers for the Curation Statistics table. So, for AFP Positives for example, the following logic is performed to determine the indicated values (list of papers), per datatype:
AFP positive (%afpPos) AFP positive validated (%afpPosVal) : %afpPos AND (%valNeg OR %valPos) AFP positive validated false positive (%afpPosFP) : %afpPos AND %valNeg AFP positive validated true positive (%afpPosTP) : %afpPos AND %valPos AFP positive validated true positive curated (%afpPosTpCur) : %afpPos AND %valPos AND %valCur <Note: the %valPOS is redundant> AFP positive validated true positive not curated (%afpPosTpNC) : %afpPos AND (%valPos NOT %valCur) AFP positive not validated (%afpPosNV) : %afpPos NOT (%valNeg OR %valPos) AFP positive not curated (%afpPosNC) : (%afpPos AND (%valPos NOT %valCur)) OR (%afpPos NOT (%valNeg OR %valPos))
and for AFP Negatives:
AFP negative (%afpNeg) AFP negative validated (%afpNegVal) : %afpNeg AND (%valNeg OR %valPos) AFP negative validated true negative (%afpNegTN) : %afpNeg AND %valNeg AFP negative validated false negative (%afpNegFN) : %afpNeg AND %valPos AFP negative validated false negative curated (%afpNegFnCur) : %afpNeg AND %valPos AND %valCur <Note: the %valPOS is redundant> AFP negative validated false negative not curated (%afpNegFnNC) : %afpNeg AND %valPos NOT %valCur AFP negative not validated (%afpNegNV) : %afpNeg NOT (%valNeg OR %valPos) AFP negative not curated (%afpNegNC) : (%afpNeg AND (%valPos NOT %valCur)) OR (%afpNeg NOT (%valPos OR %valNeg))
To determine the "Any" and "Intersection" results, all flagging methods currently visible in the Curation Statistics table are considered. So, for the main Curation Statistics table (with no options selected), all flagging methods (SVM, AFP, and CFP as of 12-10-2012) are considered. The calculations in this case would be:
Any flagged : %svmData OR %afpFlagged OR %cfpFlagged Any positive : %svmPos OR %afpPos OR %cfpPos Any positive validated : %svmPosVal OR %afpPosVal OR %cfpPosVal Any positive validated false positive : %svmPosFP OR %afpPosFP OR %cfpPosFP Any positive validated true positive : %svmPosTP OR %afpPosTP OR %cfpPosTP Any positive validated true positive curated : %svmPosTpCur OR %afpPosTpCur OR %cfpPosTpCur Any positive validated true positive not curated : %svmPosTpNC OR %afpPosTpNC OR %cfpPosTpNC Any positive not validated : %svmPosNV OR %afpPosNV OR %cfpPosNV Any positive not curated : %svmPosNC OR %afpPosNC OR %cfpPosNC Intersection flagged : %svmData AND %afpFlagged AND %cfpFlagged Intersection positive : %svmPos AND %afpPos AND %cfpPos Intersection positive validated : %svmPosVal AND %afpPosVal AND %cfpPosVal Intersection positive validated false positive : %svmPosFP AND %afpPosFP AND %cfpPosFP Intersection positive validated true positive : %svmPosTP AND %afpPosTP AND %cfpPosTP Intersection positive validated true positive curated : %svmPosTpCur AND %afpPosTpCur AND %cfpPosTpCur Intersection positive validated true positive not curated : %svmPosTpNC AND %afpPosTpNC AND %cfpPosTpNC Intersection positive not validated : %svmPosNV AND %afpPosNV AND %cfpPosNV Intersection positive not curated : %svmPosNC AND %afpPosNC AND %cfpPosNC
Note that if a curator enters the Curation Statistics table after entering deselecting any of the flagging methods in the Curation Statistics Options Page, the "Any" and "Intersection" sections of the table will only reflect the flagging methods chosen by the curator. Thus, if a curator chooses to view only one flagging method, the "Any", "Intersection", and "Flagged Positive" sections of the table will show identical results.
The following are the correspondences between rows in the Curation Statistics table and the hash tables in the form's code:
General paper stats
%curatablePapers curatable papers %objsCurated objects curated %objsCurated/%valCur objects curated per paper %valCur Papers curated %validated Papers validated %valPos Papers validated positive %valCur Papers validated positive curated %valPos NOT %valCur Papers validated positive not curated %valNeg Papers validated negative %conflict Papers validated conflict
Support Vector Machine paper stats
%noSvm SVM no svm processed %svmData SVM has svm %svmPos SVM positive any %svmPosVal SVM positive any validated %svmPosFP SVM positive any validated false positive %svmPosTP SVM positive any validated true positive %svmPosTpCur SVM positive any validated true positive curated %svmPosTpNC SVM positive any validated true positive not curated %svmPosNV SVM positive any not validated %svmPosNC SVM positive any not curated %svmHig SVM positive high %svmHigVal SVM positive high validated %svmHigFP SVM positive high validated false positive %svmHigTP SVM positive high validated true positive %svmHigTpCur SVM positive high validated true positive curated %svmHigTpNC SVM positive high validated true positive not curated %svmHigNV SVM positive high not validated %svmHigNC SVM positive high not curated %svmMed SVM positive medium %svmMedVal SVM positive medium validated %svmMedFP SVM positive medium validated false positive %svmMedTP SVM positive medium validated true positive %svmMedTpCur SVM positive medium validated true positive curated %svmMedTpNC SVM positive medium validated true positive not curated %svmMedNV SVM positive medium not validated %svmMedNC SVM positive medium not curated %svmLow SVM positive low %svmLowVal SVM positive low validated %svmLowFP SVM positive low validated false positive %svmLowTP SVM positive low validated true positive %svmLowTpCur SVM positive low validated true positive curated %svmLowTpNC SVM positive low validated true positive not curated %svmLowNV SVM positive low not validated %svmLowNC SVM positive low not curated %svmNeg SVM negative %svmNegVal SVM negative validated %svmNegTN SVM negative validated true negative %svmNegFN SVM negative validated false negative %svmNegFnCur SVM negative validated false negative curated %svmNegFnNC SVM negative validated false negative not curated %svmNegNV SVM negative not validated %svmNegNC SVM negative not curated
Author First Pass paper stats
%afpEmailed AFP emailed %afpFlagged AFP flagged %afpPos AFP positive %afpPosVal AFP positive validated %afpPosFP AFP positive validated false positive %afpPosTP AFP positive validated true positive %afpPosTpCur AFP positive validated true positive curated %afpPosTpNC AFP positive validated true positive not curated %afpPosNV AFP positive not validated %afpPosNC AFP positive not curated %afpNeg AFP negative %afpNegVal AFP negative validated %afpNegTN AFP negative validated true negative %afpNegFN AFP negative validated false negative %afpNegFnCur AFP negative validated false negative curated %afpNegFnNC AFP negative validated false negative not curated %afpNegNV AFP negative not validated %afpNegNC AFP negative not curated
Curator First Pass paper stats
%cfpFlagged CFP flagged %cfpPos CFP positive %cfpPosVal CFP positive validated %cfpPosFP CFP positive validated false positive %cfpPosTP CFP positive validated true positive %cfpPosTpCur CFP positive validated true positive curated %cfpPosTpNC CFP positive validated true positive not curated %cfpPosNV CFP positive not validated %cfpPosNC CFP positive not curated %cfpNeg CFP negative %cfpNegVal CFP negative validated %cfpNegTN CFP negative validated true negative %cfpNegFN CFP negative validated false negative %cfpNegFnCur CFP negative validated false negative curated %cfpNegFnNC CFP negative validated false negative not curated %cfpNegNV CFP negative not validated %cfpNegNC CFP negative not curated
Abbreviations
AFP - Author First Pass (flagging method)
CFP - Curator First Pass (flagging method)
OA - Ontology Annotator (curation tool)
SVM - Support Vector Machine (flagging method)
Definitions
datatype - A type of data of that WormBase curates
flagged - Processed by a flagging method (flagged positive OR negative)
flagging method - Manual or automated method for identifying research articles that contain a particular datatype
validated - Definitively confirmed by a curator to have (or not have) the relevant datatype