Pictures
links to relevant pages
Caltech documentation
PIctures
Contents
Picture Curation
The immediate goal of picture curation is to be able to obtain images of gene expression data from the literature and individual laboratories and display them in the WormBase gene expression page.
- We want display images related to the temporal or spatial (e.g., tissue, subcellular, etc.) localization of any gene in a wild-type background with different data types
- Reporter gene analysis
- Antibody staining
- In situ hybridization
- RT-PCR
- Western or Northern blot data
Pipeline
In the early phases of curation, pictures will be taken from open access journals (e.g. PLoS). During the process of PLoS image curation, other publishers will be contacted for obtaining copyright permissions.
The images should be saved and stored according to the following guidelines. The example shown below refers to a PLoS Biology paper but the rules of handling the pictures are universal and not "paper specific".
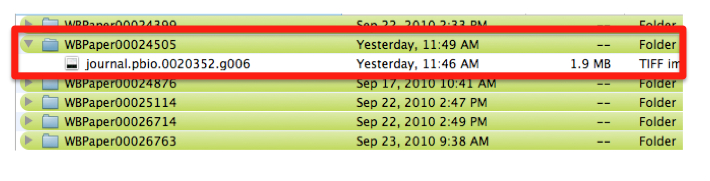
Pictures are downloaded in TIFF format from the original paper.
Pictures are saved with their original name in order to minimize editing from the curator. In this case the file is called “journal.pbio.0020352.g006”.
The file is saved in a directory named after the WB paper ID. E.g.: WBPaper00024505, meaning that picture “journal.pbio.0020352.g006” has been downloaded from WBPaper00024505.
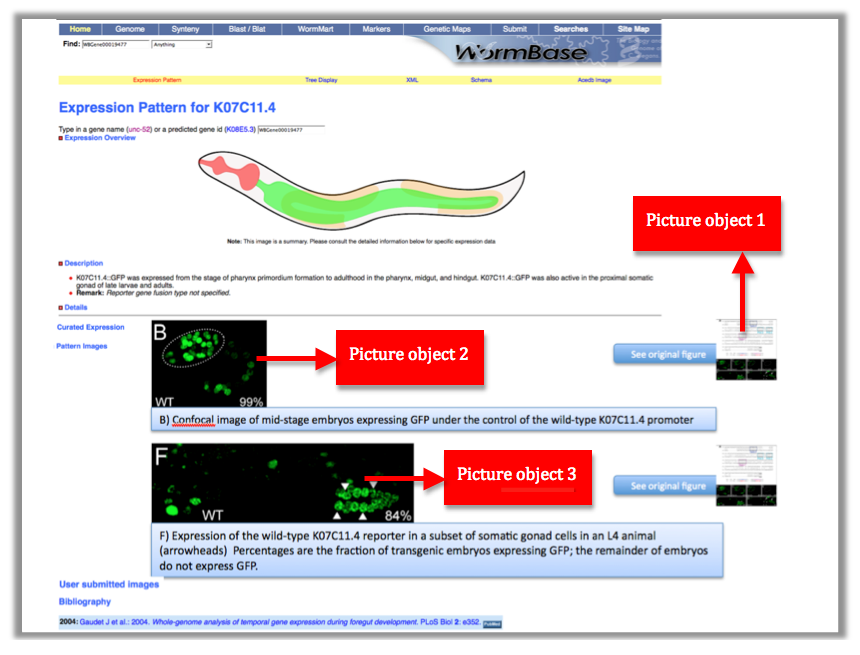
These 2 numbers together WBPaper00024505_journal.pbio.0020352.g006 will be UNIQUE IDENTIFIERS of the object, that we call Picture object 1 (WBPicture000000001). The ID WBPicture000000001 will be the NAME of the object in the Picture Data Model.
The path WBPaper00024505_journal.pbio.0020352.g006 will define the SOURCE of the object in the Picture Data Model.
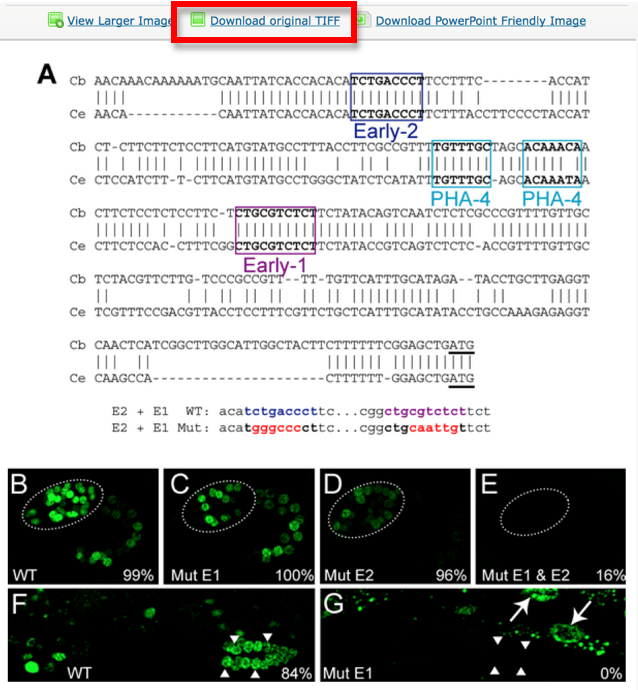
Now look at the picture above: In our WormBase expression pattern page we don’t want to display the whole picture because it contains information not pertinent to the expression data. We therefore need to CROP the 2 pictures depicting expression of the gene in the Wild Type. We want to have only panel B and F.
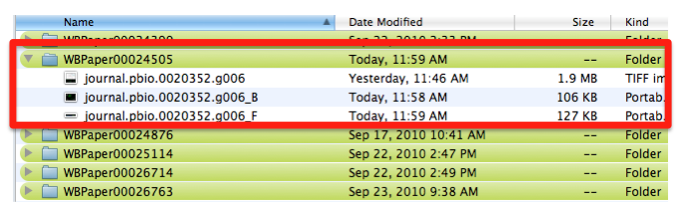
Each panel is cropped from the original picture in Photoshop and the files are saved as “journal.pbio.0020352.g006_B” “journal.pbio.0020352.g006_F” in the same directory as before: WBPaper00024505
These will be respectively Picture object 2(WBPicture000000002) and Picture object 3 (WBPicture000000003).
To summarize till now:
Picture object 1: WBPicture000000001: WBPaper00024505_ journal.pbio.0020352.g006
Picture object 2 WBPicture000000002: WBPaper00024505_ journal.pbio.0020352.g006_B
Picture object 3: WBPicture000000003: WBPaper00024505_ journal.pbio.0020352.g006_F
where WBPicture000000001 corresponds to the NAME of the object in the picture data model and WBPaper00024505_ journal.pbio.0020352.g006 corresponds to the SOURCE of the object in the picture data model.
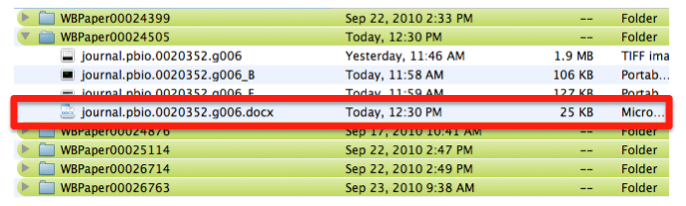
At the same time, the text file associated with the entire figure WBPicture000000001, is saved with the same name as the figure -journal.pbio.0020352.g006- with a .doc extension. In this way we can make sure which figure legend goes with which picture.
NB: The size of the original TIFF file varies from 1 to 20 MB (in the examples observed till now). In order to minimize the space, the cropped images will be converted into PNG files (100KB to 2MB). We anyway have the parental image for users access.
Supplementary material is generally stored by journals as PDF files. The PDF is converted in TIFF for consistency.
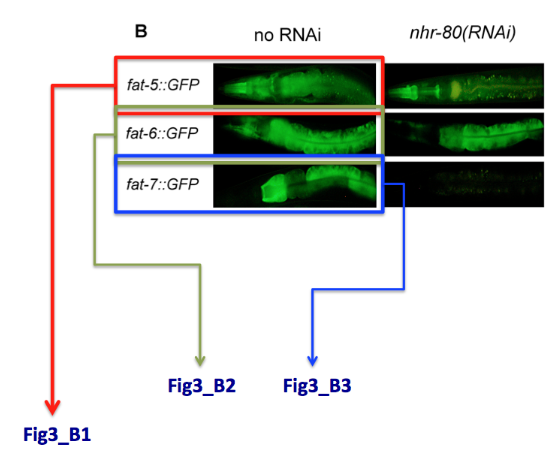
Special case: what do I do when one single panel refers to multiple genes. E.g. panel B displays the expression of 3 different genes. We will simply name the pictures Fig3_B1, Fig3_B2, Fig3_B3, see example below.
Let's go one step further:
Picture object 1 is our PARENTAL IMAGE, we will display it only when the user will click on a “see original figure” link. Picture Objects 2 and 3 are our Daughter Images, which will be displayed on the gene expression page. See mock page below for a visual example:
Picture Data Model Proposal
////////////////////////////////////////////////////////////////////////////////////
?Picture Description ?Text
Name ?Text
Source ?Text
Image_lineage Crop_picture ?Picture XREF Cropped_from
Cropped_from ?Picture XREF Crop_picture
Pick_me_to_call Text Text
Expr_pattern ?Expr_pattern XREF Picture
RNAi ?RNAi XREF Picture
Variation ?Variation XREF Picture
Transgene ?Transgene XREF Picture
Reference ? XREF Picture
Remark ?Text #Evidence
///////////////////////////////////////////////////////////////////////////////////
Picture Data Model step by step explanation
Description
Figure legend
Name
Name of the picture object e.g. WBPicture0000000001
Source
For actual picture names. This is the name of the path leading to the picture file. The source includes the name of the directory where the picture comes from AND the name of the picture file. e.g. WBPaper00024505_journal.pbio.0020352.g006
Image Lineage
This is the picture object lineage. Large figures will be cropped into sections when they represent different data. We want to maintain the picture lineage -> by clicking on the "see original figure button" we want to access the entire image.
Pick_me_to_call
It is probably an XACE command to call the image. We leave it there for now and ask to the web team if it is necessary