Co-op Documentation
Contents
- 1 Current Tasks
- 1.1 Tasks In Progress
- 1.1.1 General Page Corrections
- 1.1.2 Refining Interactions widget
- 1.1.3 Gene page alterations
- 1.1.4 Sequence/Transcript/CDS page alterations
- 1.1.5 Variation page alterations
- 1.1.6 Curator Requests
- 1.1.7 Non-compliant Data Checks
- 1.1.8 Interface Corrections
- 1.1.9 Homology Widget
- 1.1.10 Cytoscape plugin
- 1.2 Other Tasks
- 1.1 Tasks In Progress
- 2 Getting Started
- 3 Tools/Resources
- 4 General Info
- 5 General Concepts
- 6 Other Info
Current Tasks
Tasks In Progress
General Page Corrections
- Details:
- Visiting all minor pages and verifying that all fields display information correctly (checking behaviour for sparsely- & densely-populated)
- Continuing with fatal non-compliance checks for all pages
- Progress:
- Related/Example Links:
- Completed:
Refining Interactions widget
- Details:
- Scale distance based from current gene based on connectedness(number of interactions)
- Shouldn't cluster by gene class (problems arise from classes like "let" where genes aren't necessarily related, so grouping/clustering can be misleading)
- Interesting example where this seems to happen automatically: pptr-1
- Shouldn't cluster by gene class (problems arise from classes like "let" where genes aren't necessarily related, so grouping/clustering can be misleading)
- Scale distance based from current gene based on connectedness(number of interactions)
- Progress:
- Checking cytoscapeweb documentation for how to incorporate this
- Related/Example Links:
- unc-26(
8271) - daf-16(
433181) - old daf-16 (beta)(1386 w/ duplication)
- daf-2(600 no nearby)
- unc-26(
- Completed:
- If the number of interactions is too high, only show the minimal table (hide Cytoscape?) and add "Show More"/"Show Neighbouring Interactions"? option
- Otherwise, show all interactions(nearby interactions included) and Cytoscape plugin
- NOTE: This may need to be revisited and optimized/cleaned up
- Looking into BLASTP field in Homology Widget on Protein page that uses similar mechanism
- remove duplication
- instead of toggle href load, load diagram with no nearby in toggle
- create threshold-determining algorithm based on nodes?
- Split interactions into 2 methods, one for those involving current gene and one for nearby interactions
- If local interactions exceeds
400100(after merging duplicates), doesn't load nearby - Combined duplicate edges (duplicate implies matching source node, target node, direction, type, & phenotype)
- Changed table: "Details" column with single link to interaction object -> "Citations" column with list of links to duplicate interactions
- More than 10 results in a cell is now collapsible into "# Results"
- Modified tooltip
- Removed interaction number and link to page (misleading for multiple)
- Added phenotype and number of citations to tooltip
- Added "edge width based on number of citations" feature
- Set 15 as max number of citations; anything higher will be scaled down
- Change colour scheme to follow WormBase's
- Start by looking in main.css
- If the number of interactions is too high, only show the minimal table (hide Cytoscape?) and add "Show More"/"Show Neighbouring Interactions"? option
Gene page alterations
- Details:
- Making corrections to Gene page interface based on curator consultation
- Progress:
- Interactions info needs to be loaded from a separate database to speed up
- Related/Example Links:
- Completed:
- Phenotype:
- Added evidence information
- May need to be modified/re-organized (pending consultation)
- Gene Ontology widget uses similar mechanism
- _get_evidence method in Object.pm
- Added evidence information
- Consultation from curators. Teleconference: 11:30 am EST Wednesday, January 18th, 2012
- Interactions:
- Added edge-colour legend
- Hide no_interaction edges unless that option is selected/filtered for by user
- Phenotype:
- Standardize look and feel of tables (convert to data tables)
- Done changing Phenotypes, Phenotypes_not_observed, and RNAi tables to use build_data_table
- Modified corresponding API methods to return compatible data for build_data_table
- Combined/merged Phenotype and RNAi tables for observed and not_observed (with >10 results collapsible)
- Standardize look and feel of tables (convert to data tables)
- Reagents:
- Drop the word "found" for search results
- Switch search results to actual list of results(collapsible if too many)
- See/learn how this was done for the Expression widget
- implemented using tags2link macro
- Homology:
- restore best blast hits (on the protein object)
- protein domains: alphabetize & restrict to just interpro hits
- Changing evidence in Phenotypes widget to be consistent
- Format: "name/id(paper)" with evidence drop-down when clicked for all types - Alleles, Transgenes,& RNAi(extract info)
- Phenotype:
Sequence/Transcript/CDS page alterations
- Details:
- Making corrections to Sequence, CDS, & Transcript pages based on curator consultation
- CDS
- Sequences Widget
- Show marked-up sequences for corresponding transcripts
- Sequences Widget
- All
- Overview Widget
- Download dialog launched from highlight box: fix scrollbar and rapid-click issues
- Location Widget
- Highlight current on track or at least show gene names
- Overview Widget
- Progress:
- Split CDS and Transcript, now checking that methods do not attempt to access invalid fields
- Related/Example Links:
- JC8.10c (CDS)
- JC8.10c.1 (Transcript)
- yk444b8.5 (Sequence)
- Completed:
- Consultation from curators. Teleconference: 11:30 am EST Wednesday, January 25th, 2012
- Separating Transcript(+CDS) and Sequence pages
- Separating CDS from Transcript
- Checking Sequence page methods to ensure that only valid fields are accessed
- Comparing with schema
- Removing eval statements
- Region Widget
- Redistribute data and remove widget
- Matching cDNAs moved to Reagents widget
- Moved "Transcripts in this region" field to Sequences widget
- Redistribute data and remove widget
- Origin Widget
- Moved to Overview
- Changed to a curator-only field using curator_block macro
- Reagents Widget
- Add PCR products
- Tags/info should already be in the model
- Add PCR products
- Sequences Widget
- Predicted Exon Structure table
- Removed completely (Although tag exists in schema, it has not been populated for any Sequences, only Transcripts)
Calculate and show exon & intron sizes using "Relative to Itself"Remove "Relative to Superlink"Start & End "Relative to Itself" are useful in some cases (keep)
- Predicted Exon Structure table
- Sequences
- Sequences Widget
- Show orthologs? (for navigation to related info, especially useful for curators)
- Syntenic sequences
- Show corresponding gene in predicted genes and transcriptional units (e.g. unc-26 for yk444b8.5)
- Show orthologs? (for navigation to related info, especially useful for curators)
- Sequences Widget
- Transcript
- Split into CDS and Transcript
- Location Widget
- Show protein domains for CDS
- All
- Overview Widget
- Make origin info available to users (not just curators)
- Show remarks/description associated with Method to make it more meaningful
- e.g. Method for AAA27985.1 is ndb_cds which is not particularly meaningful
- Show affiliated gene(e.g. unc-26 for yk444b8.5)
- Add navigation table (similar to table in sequences widget on gene page, e.g. Matching transcripts/CDS)
- Location Widget
- Hide YAC's, fosmids, & cosmids track
- Overview Widget
Variation page alterations
- Details:
- Making corrections to Variation page interface based on curator consultation
- Change default widgets (Overview, Genetics, Molecular Details, Phenotypes)
- Overview Widget
- Hide Status field if Live (show otherwise e.g. suppressed/dead)
- Remove redundant remarks
- Molecular Details Widget
- Check that the sequence is actually on the + strand?
- "Flanking Sequences" and "Context" should be removed
- Just show marked-up sequence by default (toggle if too large)
- Add colour legend (yellow=flanking, red=deletion/substitution/...)
- Features Affected field -> Predicted CDSs
- Add "Variation" to "Contained in:" to clarify
- Change Predicted CDSs and Clone to data tables
- Location Widget
- Remove contig submission track
- Phenotypes
- Remove remarks and add evidence/citations
- Remove phenotype description or hide by default
- Progress:
- Related/Example Links:
- Completed:
- Consultation from curators. Teleconference: 11:30 am EST Wednesday, February 1st, 2012
Curator Requests
- Details:
- Protein Page (Christian A Grove)
- UNC-26 (Sample Protein Page)
- Overview widget: in the right-hand box, the "Status: history" is not clear. What does this mean? Do we need/want to display this?
- External Links widget: "TREEFAM" links back to the same UNC-26 protein page, as does "WORMPEP". "WP:CE28239" links to the old UNC-26 protein page; is this what we want? Also, both UniProt links are dead/obsolete/outdated
- We should add units to Molecular Weight wherever it is mentioned (as I said in the gene page review). The units should either be "kilodaltons" or "kD"
- History widget: The "JC8.10" under "Predicted gene" links to a WormBase search result for "JC8.10". I think it would be better to direct users to the exact (unc-26) page, rather than having them have to select among many options.
- The red text in the protein schematic for domain names is a bit hard on the eyes. Could we go with a more neutral color: black, grey, blue...?
- Can we include descriptions of the domains depicted in the domain cartoons, or at least provide a link out to the relevant InterPro or PFAM pages?
- Gene Page (Christian A Grove)
- cup-5 (Sample Gene Page)
- Postponed:
- Human Diseases widget:
- The description for the disease is cutoff: "...along the lysosomal pathway, affecting membra" ; is this intentional? Could we maybe add a "..." to the end?
- Phenotype widget
- Cross-platform issues (Chrome & Safari vs Firefox)
- The table display for "Primary Sequence ID" in the Sequences widget is a little wonky on Firefox. As you can see from the screenshots (attached) as I resize the table in Firefox, that column adjusts strangely, skewing the text, whereas in Safari and Chrome, the table stays nice and neat.
- Human Diseases widget:
- There are a number of unnecessary hyperlinks in the phenotype description details. For example, all remarks, the word "Genotype" after "Phenotype assay:", and the phrase "Uncharacterised_loss_of_function" after "Loss of function:" are all hyperlinked to what appears to be a general WormBase text search result. These should just be plain text.
- One general comment is that I think I would prefer that links out to other pages open a new tab by default, but I may be in the minority with that opinion (or that may have been your intention, but Firefox is not cooperating)
- Protein Page (Christian A Grove)
- Progress:
- Investigating hyperlinks in phenotype description details
- Related/Example Links:
- Completed:
- Gene Page
- External links widget:
- The OMIM link out appears to be malformed. The URL that the link directs me too is:
- http://omim.org/enrty/OMIM:605248 (verbatim, entry is mispelled)
- whereas this is a working link:
- (as I write this I realize that someone from OMIM just e-mailed to point this out)
- Queried ACeDB Database class for databases with URL, URL_Constructor, and Description and added those to the external_urls template
- Need to verify that all links are working and add other links(i.e. databases that didn't match the query)
- The OMIM link out appears to be malformed. The URL that the link directs me too is:
- Homology widget:
- When I click on the download option for homologous proteins, a window pops up to show Sequence, Isoelectric Point, Molecular Weight, etc. I know this is more a Protein issue, but we should put units for Molecular Weight (e.g. "77.7 kiloDaltons" or "kD" as opposed to just "77.7")
- Genetics widget:
- In both the "Alleles" and the "Polymorphisms & Natural Variants" tables, the right-most column is "Protein effect" and "Location", respectively. Under "Protein effect" is displayed "nonsense" and "missense" (which are OK), but also "intron", which does not make sense. Under "Location" is displayed "intron" and "utr_3" (OK), but also "missense" and "silent" (not OK). I would suggest having a "Protein effect" and a "Location" column in each table and putting only relevant entries in each:
- Protein effect: nonsense, missense, silent, etc.
- Location: intron, exon, utr_3, utr_5, etc.
- In both the "Alleles" and the "Polymorphisms & Natural Variants" tables, the right-most column is "Protein effect" and "Location", respectively. Under "Protein effect" is displayed "nonsense" and "missense" (which are OK), but also "intron", which does not make sense. Under "Location" is displayed "intron" and "utr_3" (OK), but also "missense" and "silent" (not OK). I would suggest having a "Protein effect" and a "Location" column in each table and putting only relevant entries in each:
- Interactions widget:
- The different color nodes and thickness of lines are not explained in the legend. Can we add an explanation somewhere?
- In the "Citations" column is listed all of the WormBase Interaction objects affiliated with this interaction. I would expect to see paper references in a "Citations" column. Could we perhaps put the interactions under a "Interactions" column all the way to the left, and keep the "Citations" column at the right with WBPaper references?
- The Cytoscape network view looks great, but the window for viewing it is too short (vertically) in Firefox, and too tall in Safari and Chrome. Could we find an intermediate height or provide options to resize?
- External links widget:
- Gene Page
Non-compliant Data Checks
- Details:
- Using fatal_non_compliance check and debug option (enable in wormbase_local.conf) to determine if API methods return invalid data
- E.g. returning ACe objects(usually this should be a string) or empty arrays/hashes(should return undef)
- Usually these can be found when fields do not have data to return but give empty structures instead of just undef
- Using fatal_non_compliance check and debug option (enable in wormbase_local.conf) to determine if API methods return invalid data
- Progress:
- Correcting these as they're encountered
- Completed:
- Methods showing errors on the following pages were corrected:
- Variation, Person, Laboratory, Gene Class, Life Stages, Phenotype, Protein, Strain
- Methods showing errors on the following pages were corrected:
Interface Corrections
- Details:
- Look into rearranging the elements in widgets so that they do not appear strange/distorted when switched from one- to two-column views or using smaller displays
- Will require modifying the templates
Homology Widget
- Needs to be modified; Contains too much information that shouldn't necessarily be grouped this way
- May need to move orthologs to separate widget
Cytoscape plugin
- Incorporate button/option to open pop-up for larger view (e.g. in case user has small screen or is using 2-column layout)
- Also, add plugin to interaction page (may need to wait until interactions are merged)
Other Tasks
Additional tasks that require further investigation
- Loading/processing large amounts of data
- Details:
- Likely approach will be to change the database used from ACeDB to NoSQL/CouchDB
- Sample Issue
- Progress:
- Ace2Couch Scripts for migrating data
- AceCouch Perl API
- Details:
Getting Started
Members/Contacts
- Abigail Cabunoc - abigail.cabunoc@oicr.on.ca
- Todd Harris - todd@wormbase.org
- Lincoln Stein - lincoln.stein@gmail.com
- Quang Trinh
Meetings
- WormBase OICR Developers Teleconference
- Mondays, 3:00PM
- Phone-in 1-800-747-5150 id: 6738514
- WormBase OICR Developers Teleconference with Lincoln
- Wednesdays, 4:00PM EST
- Lincoln's office
- Agenda and Minutes
- WormBase International Groups Teleconference
- Every other Thursday, 11:30AM
- Lincoln's Office
- Alternatively: 1-866-528-2256
- Access Code: 714646
- Group Meeting
- Fridays, 3:00PM
- HL31 Conference Room/TBA
Tools/Resources
The following are some of the tools with which many major aspects of WormBase are developed. You should familiarize yourself with them through documentation and examples.
Perl
There is significant documentation on getting started with Perl. One starting point is PerlMonks For information regarding the use of ACeDB in conjuction with Perl (e.g. retrieving data), check AcePerl Documentation. The Ace::Object section contains most of the information related to interacting with ACeDB objects.
Catalyst
Catalyst is the web development framework used to develop WormBase. To get started, read and try examples from
- The Definitive Guide to Catalyst (should be available on bookshelf)
- CPAN - About Catalyst
Git
Git is a version control system used for collaboration and backup in the development process. One starting point is the progit tutorial
- WormBase repository located at https://github.com/organizations/WormBase
- Common commands
- status, add, checkout, commit, push, pull, log, fetch, merge
- Usage example:
Given that we have modified two files a.txt and b.txt but do not wish to keep the changes made to b.txt
git status
git checkout b.txt
git add a.txt
git commit -m “Added change1 and change2 to a.txt”
git push
If we have did not have the most recent version, then we will run into an issue when trying to push. In this case we can:
git fetch
git merge
git push
Note: pull is similar to using fetch + merge
Browser (Debugging)
Many browsers provide useful tools to developers that can be used for debugging
Chrome
- Tools->Developer Tools/JavaScript Console
Firefox
- Firefox->Web Developer->Web Console/Error Console
Other
Some other tools that you should be aware of but may not be required to know/interact with include:
- JavaScript
- ACeDB
- Usage:
- Navigate to /usr/local/wormbase/acedb/bin and run: "./tace ../wormbase"
- For more help, try the tace tutorial
- Usage:
- MySQL
- Xapian
- GFF
- Cytoscape Web
- Cytoscape Tutorial
- Plugin generally used for pathway analysis
- This plugin is used in the Interactions widget on the Gene page ([WormBase dir]/root/templates/classes/gene/interactions.tt2)
- Installation directory (for updating): [WormBase dir]/root/js/jquery/plugins/
General Info
- Unix and Cluster training by Quang (OICR login required)
General Concepts
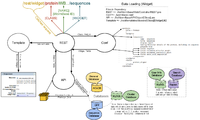
Widget Data Loading
REST Controller:
- Catches internal url (/rest/widget/...)
- Determines the class and widget from the url
- From class and widget, determines which fields are required from the configuration file (wormbase.conf)
- API methods request data from appropriate databases, process/format/package the data, and then return it(/lib/WormBase/API/Object/[Class].pm)
- Sends data to the template to be used in rendering the widget (/root/templates/classes/[Class]/[Widget].tt2)
Other Info
Useful Macros when editing templates can be found in:
- /root/templates/config/main
- e.g. tags2link
- /root/templates/shared/page_elements.tt2
- e.g. build_data_table
Useful files for debugging include:
- /logs/wb-dev-catalyst.log
Other:
- If the port you have been using appears to be busy but the server is not running,
ps -aux|grep -XXXX kill -9 ID
where XXXX is the port number(e.g. 8023) and ID is the process id that is using the port(e.g. 10361) NOTE: Do not kill other users' processes if you are using a shared dev machine (e.g. wb-dev)
- To dump data in API methods include Data::Dumper
use Data::Dumper ... warn(Dumper(\@data));
- Sometimes firewall blocks certain ports, so you may be unable to connect to your dev server on the wb-dev machine through the browser
ssh -L 8080:localhost:XXXX wb-dev.oicr.on.ca ./wormbase_server.pl -p XXXX -d -r
Then go to localhost:8080 in browser